Lectures 5 and 6: Class demo#
Imports, Announcements, LOs#
Imports#
# import the libraries
import os
import sys
sys.path.append(os.path.join(os.path.abspath("../"), "code"))
from plotting_functions import *
from utils import *
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
%matplotlib inline
pd.set_option("display.max_colwidth", 200)
Do you recall the restaurants survey you completed at the start of the course?
Let’s use that data for this demo. You’ll find a wrangled version in the course repository.
df = pd.read_csv('../data/cleaned_restaurant_data.csv')
df
| north_america | eat_out_freq | age | n_people | price | food_type | noise_level | good_server | comments | restaurant_name | target | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Yes | 3.0 | 29 | 10.0 | 120.0 | Italian | medium | Yes | Ambience | NaN | dislike |
| 1 | Yes | 2.0 | 23 | 3.0 | 20.0 | Canadian/American | no music | No | food tastes bad | NaN | dislike |
| 2 | Yes | 2.0 | 21 | 20.0 | 15.0 | Chinese | medium | Yes | bad food | NaN | dislike |
| 3 | No | 2.0 | 24 | 14.0 | 18.0 | Other | medium | No | Overall vibe on the restaurant | NaN | dislike |
| 4 | Yes | 5.0 | 23 | 30.0 | 20.0 | Chinese | medium | Yes | A bad day | NaN | dislike |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 959 | No | 10.0 | 22 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | like |
| 960 | Yes | 1.0 | 20 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | like |
| 961 | No | 1.0 | 22 | 40.0 | 50.0 | Chinese | medium | Yes | The self service sauce table is very clean and the sauces were always filled up. | Haidilao | like |
| 962 | Yes | 3.0 | 21 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | like |
| 963 | Yes | 3.0 | 27 | 20.0 | 22.0 | Other | medium | Yes | Lots of meat that was very soft and tasty. Hearty and amazing broth. Good noodle thickness and consistency | Uno Beef Noodle | like |
964 rows × 11 columns
df.describe()
| eat_out_freq | age | n_people | price | |
|---|---|---|---|---|
| count | 964.000000 | 964.000000 | 6.960000e+02 | 696.000000 |
| mean | 2.585187 | 23.975104 | 1.439254e+04 | 1472.179152 |
| std | 2.246486 | 4.556716 | 3.790481e+05 | 37903.575636 |
| min | 0.000000 | 10.000000 | -2.000000e+00 | 0.000000 |
| 25% | 1.000000 | 21.000000 | 1.000000e+01 | 18.000000 |
| 50% | 2.000000 | 22.000000 | 2.000000e+01 | 25.000000 |
| 75% | 3.000000 | 26.000000 | 3.000000e+01 | 40.000000 |
| max | 15.000000 | 46.000000 | 1.000000e+07 | 1000000.000000 |
Are there any unusual values in this data that you notice? Let’s get rid of these outliers.
upperbound_price = 200
lowerbound_people = 1
df = df[~(df['price'] > 200)]
restaurant_df = df[~(df['n_people'] < lowerbound_people)]
restaurant_df.shape
(942, 11)
restaurant_df.describe()
| eat_out_freq | age | n_people | price | |
|---|---|---|---|---|
| count | 942.000000 | 942.000000 | 674.000000 | 674.000000 |
| mean | 2.598057 | 23.992569 | 24.973294 | 34.023279 |
| std | 2.257787 | 4.582570 | 22.016660 | 29.018622 |
| min | 0.000000 | 10.000000 | 1.000000 | 0.000000 |
| 25% | 1.000000 | 21.000000 | 10.000000 | 18.000000 |
| 50% | 2.000000 | 22.000000 | 20.000000 | 25.000000 |
| 75% | 3.000000 | 26.000000 | 30.000000 | 40.000000 |
| max | 15.000000 | 46.000000 | 200.000000 | 200.000000 |
Data splitting#
We aim to predict whether a restaurant is liked or disliked.
# Separate `X` and `y`.
X = restaurant_df.drop(columns=['target'])
y = restaurant_df['target']
Below I’m perturbing this data just to demonstrate a few concepts. Don’t do it in real life.
X.at[459, 'food_type'] = 'Quebecois'
X['price'] = X['price'] * 100
# Split the data
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=123)
EDA#
X_train.hist(bins=20, figsize=(12, 8));
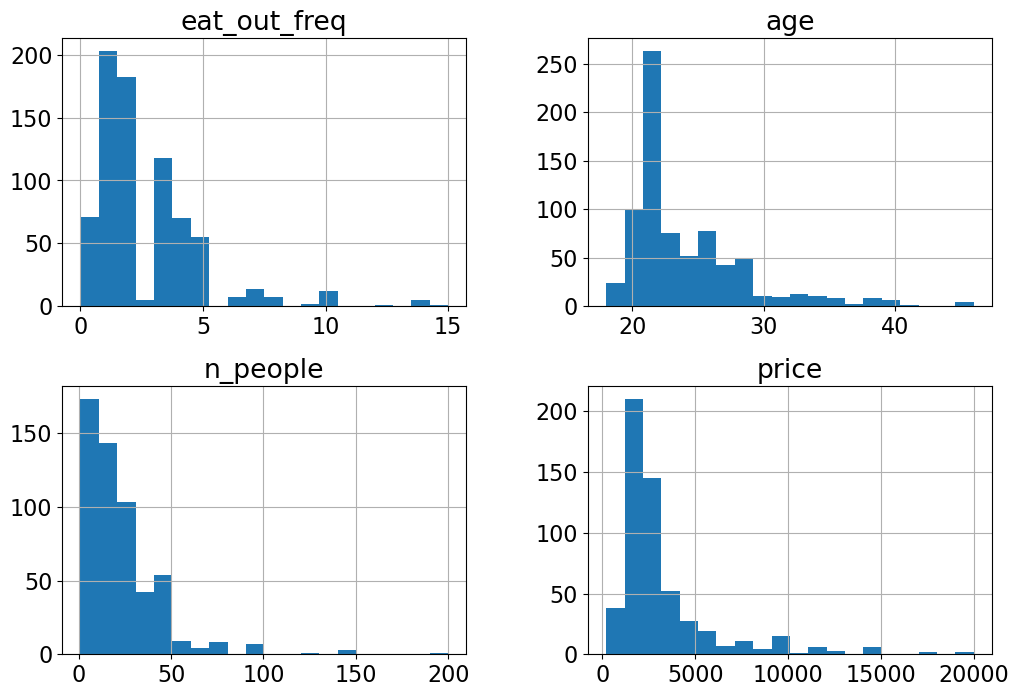
Do you see anything interesting in these plots?
X_train['food_type'].value_counts()
food_type
Other 189
Canadian/American 131
Chinese 102
Indian 36
Italian 32
Thai 20
Fusion 18
Mexican 17
fusion 3
Quebecois 1
Name: count, dtype: int64
Error in data collection? Probably “Fusion” and “fusion” categories should be combined?
X_train['food_type'] = X_train['food_type'].replace("fusion", "Fusion")
X_test['food_type'] = X_test['food_type'].replace("fusion", "Fusion")
X_train['food_type'].value_counts()
food_type
Other 189
Canadian/American 131
Chinese 102
Indian 36
Italian 32
Fusion 21
Thai 20
Mexican 17
Quebecois 1
Name: count, dtype: int64
Again, usually we should spend lots of time in EDA, but let’s stop here so that we have time to learn about transformers and pipelines.
Dummy Classifier#
from sklearn.dummy import DummyClassifier
dummy = DummyClassifier()
scores = cross_validate(dummy, X_train, y_train, return_train_score=True)
pd.DataFrame(scores)
| fit_time | score_time | test_score | train_score | |
|---|---|---|---|---|
| 0 | 0.000737 | 0.000622 | 0.516556 | 0.514950 |
| 1 | 0.000475 | 0.000337 | 0.516556 | 0.514950 |
| 2 | 0.000459 | 0.000334 | 0.516556 | 0.514950 |
| 3 | 0.000457 | 0.000330 | 0.513333 | 0.515755 |
| 4 | 0.000451 | 0.000329 | 0.513333 | 0.515755 |
We have a relatively balanced distribution of both ‘like’ and ‘dislike’ classes.
Let’s try KNN on this data#
Do you think KNN would work directly on X_train and y_train?
# Preprocessing and pipeline
from sklearn.neighbors import KNeighborsClassifier
knn = KNeighborsClassifier()
# knn.fit(X_train, y_train)
We need to preprocess the data before passing it to ML models. What are the different types of features in the data?
X_train.head()
| north_america | eat_out_freq | age | n_people | price | food_type | noise_level | good_server | comments | restaurant_name | |
|---|---|---|---|---|---|---|---|---|---|---|
| 80 | No | 2.0 | 21 | 30.0 | 2200.0 | Chinese | high | No | The environment was very not clean. The food tasted awful. | NaN |
| 934 | Yes | 4.0 | 21 | 30.0 | 3000.0 | Canadian/American | low | Yes | The building and the room gave a very comfy feeling. Immediately after sitting down it felt like we were right at home. | NaN |
| 911 | No | 4.0 | 20 | 40.0 | 2500.0 | Canadian/American | medium | Yes | I was hungry | Chambar |
| 459 | Yes | 5.0 | 21 | NaN | NaN | Quebecois | NaN | NaN | NaN | NaN |
| 62 | Yes | 2.0 | 24 | 20.0 | 3000.0 | Indian | high | Yes | bad taste | east is east |
What all transformations we need to apply before training a machine learning model?
Can we group features based on what type of transformations we would like to apply?
X_train.columns
Index(['north_america', 'eat_out_freq', 'age', 'n_people', 'price',
'food_type', 'noise_level', 'good_server', 'comments',
'restaurant_name'],
dtype='object')
X_train['noise_level'].value_counts()
noise_level
medium 232
low 186
high 75
no music 37
crazy loud 18
Name: count, dtype: int64
numeric_feats = ['age', 'n_people', 'price'] # Continuous and quantitative features
categorical_feats = ['north_america', 'food_type'] # Discrete and qualitative features
binary_feats = ['good_server'] # Categorical features with only two possible values
ordinal_feats = ['noise_level'] # Some natural ordering in the categories
noise_cats = ['no music', 'low', 'medium', 'high', 'crazy loud']
drop_feats = ['comments', 'restaurant_name'] # Let's drop them for now.
Let’s begin with numeric features. What if we just use numeric features to train a KNN model? Would it work?
# knn.fit(X_train[numeric_feats], y_train)
X_train_num = X_train[numeric_feats]
X_test_num = X_test[numeric_feats]
We need to deal with NaN values.
sklearn’s SimpleImputer#
# Impute numeric features using SimpleImputer
from sklearn.impute import SimpleImputer
imputer = SimpleImputer(strategy="median")
imputer.fit(X_train_num)
X_train_num_imp = imputer.transform(X_train_num)
X_test_num_imp = imputer.transform(X_test_num)
knn.fit(X_train_num_imp, y_train)
KNeighborsClassifier()In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook.
On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
KNeighborsClassifier()
No more errors. It worked! Let’s try cross validation.
cross_val_score(knn, X_train_num_imp, y_train).mean()
0.5418278145695364
We have slightly improved results in comparison to the dummy model.
Discussion questions#
What’s the difference between sklearn estimators and transformers?
Can you think of a better way to impute missing values?
Do we need to scale the data?
X_train[numeric_feats]
| age | n_people | price | |
|---|---|---|---|
| 80 | 21 | 30.0 | 2200.0 |
| 934 | 21 | 30.0 | 3000.0 |
| 911 | 20 | 40.0 | 2500.0 |
| 459 | 21 | NaN | NaN |
| 62 | 24 | 20.0 | 3000.0 |
| ... | ... | ... | ... |
| 106 | 27 | 10.0 | 1500.0 |
| 333 | 24 | 12.0 | 800.0 |
| 393 | 20 | 5.0 | 1500.0 |
| 376 | 20 | NaN | NaN |
| 525 | 20 | 50.0 | 3000.0 |
753 rows × 3 columns
# Scale the imputed data
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler()
scaler.fit(X_train_num_imp)
X_train_num_imp_scaled = scaler.transform(X_train_num_imp)
X_test_num_imp_scaled = scaler.transform(X_test_num_imp)
What are some alternative methods for scaling?#
MinMaxScaler: Transform each feature to a desired range
RobustScaler: Scale features using median and quantiles. Robust to outliers.
Normalizer: Works on rows rather than columns. Normalize examples individually to unit norm.
MaxAbsScaler: A scaler that scales each feature by its maximum absolute value.
What would happen when you apply
StandardScalerto sparse data?
You can also apply custom scaling on columns using
FunctionTransformer. For example, when a column follows the power law distribution (a handful of your values have many data points whereas most other values have few data points) log scaling is helpful.
For now, let’s focus on
StandardScaler. Let’s carry out cross-validation
cross_val_score(knn, X_train_num_imp_scaled, y_train)
array([0.55629139, 0.49006623, 0.56953642, 0.54 , 0.53333333])
In this case, we don’t see a big difference with StandardScaler. But usually, scaling is a good idea.
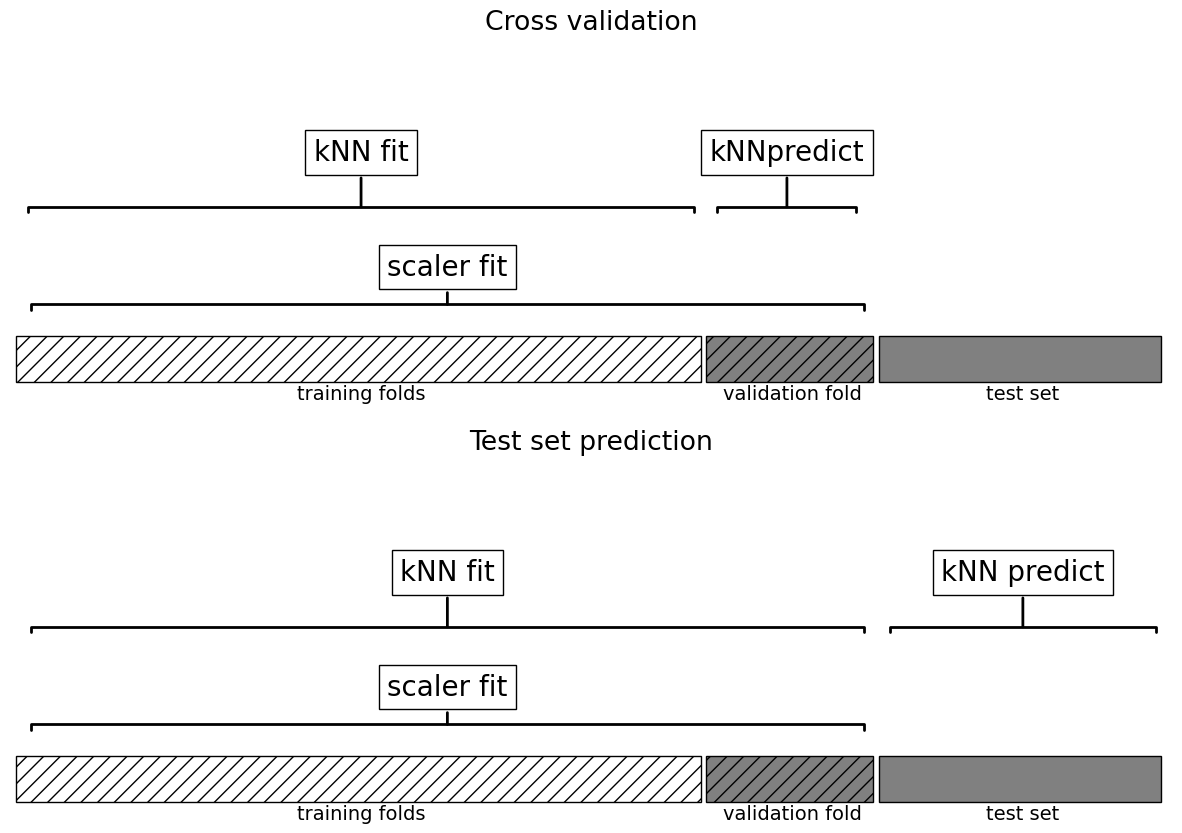
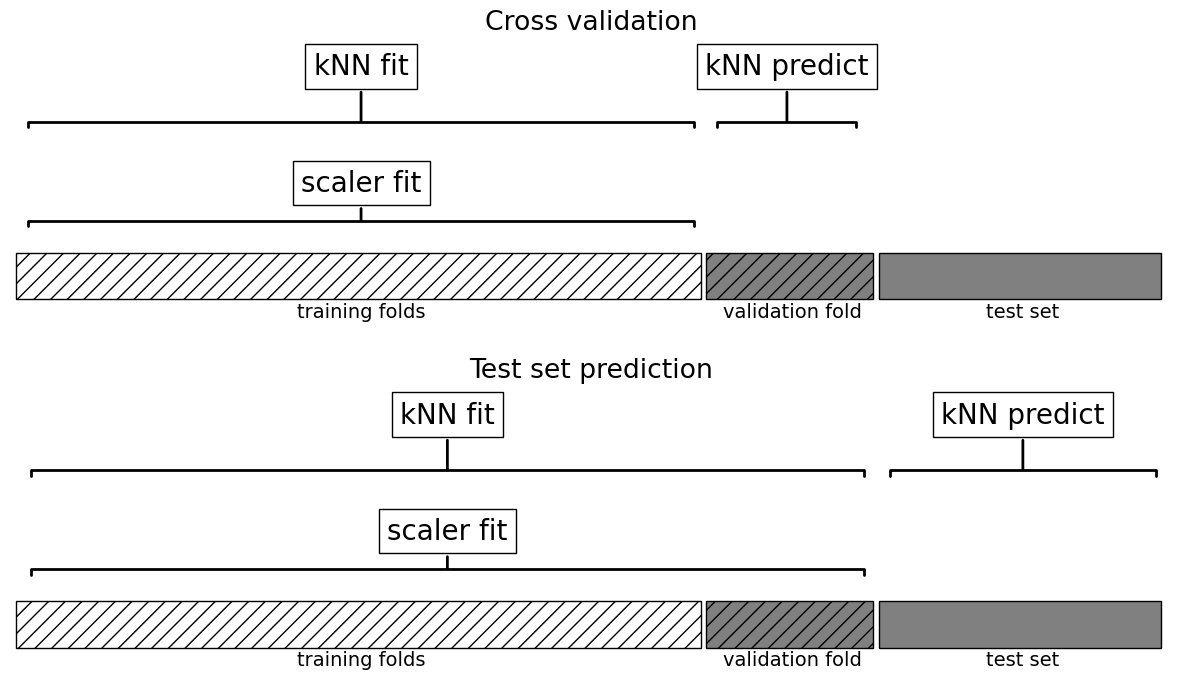
This worked but are we doing anything wrong here?
What’s the problem with calling
cross_val_scorewith preprocessed data?How would you do it properly?
# Create a pipeline
pipe_knn = make_pipeline(
SimpleImputer(strategy = "median"),
StandardScaler(),
KNeighborsClassifier()
)
cross_val_score(pipe_knn, X_train_num, y_train).mean()
0.5245916114790287
What all things are happening under the hood?
Why is this a better approach?
plot_improper_processing("kNN")
plot_proper_processing("kNN")
Categorical features#
Let’s assess the scores using categorical features.
X_train['food_type'].value_counts()
food_type
Other 189
Canadian/American 131
Chinese 102
Indian 36
Italian 32
Fusion 21
Thai 20
Mexican 17
Quebecois 1
Name: count, dtype: int64
X_train[categorical_feats]
| north_america | food_type | |
|---|---|---|
| 80 | No | Chinese |
| 934 | Yes | Canadian/American |
| 911 | No | Canadian/American |
| 459 | Yes | Quebecois |
| 62 | Yes | Indian |
| ... | ... | ... |
| 106 | No | Chinese |
| 333 | No | Other |
| 393 | Yes | Canadian/American |
| 376 | Yes | NaN |
| 525 | Don't want to share | Chinese |
753 rows × 2 columns
X_train['north_america'].value_counts()
north_america
Yes 415
No 330
Don't want to share 8
Name: count, dtype: int64
X_train['food_type'].value_counts()
food_type
Other 189
Canadian/American 131
Chinese 102
Indian 36
Italian 32
Fusion 21
Thai 20
Mexican 17
Quebecois 1
Name: count, dtype: int64
X_train_cat = X_train[categorical_feats]
X_test_cat = X_test[categorical_feats]
# One-hot encoding of categorical features
from sklearn.preprocessing import OneHotEncoder
# Define and fit OneHotEncoder
ohe = OneHotEncoder(sparse_output=False)
ohe.fit(X_train_cat)
X_train_cat_ohe = ohe.transform(X_train_cat) # transform the train set
X_test_cat_ohe = ohe.transform(X_test_cat) # transform the test set
X_train_cat_ohe
array([[0., 1., 0., ..., 0., 0., 0.],
[0., 0., 1., ..., 0., 0., 0.],
[0., 1., 0., ..., 0., 0., 0.],
...,
[0., 0., 1., ..., 0., 0., 0.],
[0., 0., 1., ..., 0., 0., 1.],
[1., 0., 0., ..., 0., 0., 0.]])
It’s a sparse matrix.
Why? What would happen if we pass
sparse_output=False? Why we might want to do that?
# Get the OHE feature names
ohe_feats = ohe.get_feature_names_out(categorical_feats).tolist()
pd.DataFrame(X_train_cat_ohe, columns = ohe_feats)
| north_america_Don't want to share | north_america_No | north_america_Yes | food_type_Canadian/American | food_type_Chinese | food_type_Fusion | food_type_Indian | food_type_Italian | food_type_Mexican | food_type_Other | food_type_Quebecois | food_type_Thai | food_type_nan | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.0 | 1.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 1 | 0.0 | 0.0 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 2 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 3 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 |
| 4 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 748 | 0.0 | 1.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 749 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 |
| 750 | 0.0 | 0.0 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 751 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 |
| 752 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
753 rows × 13 columns
cross_val_score(knn, X_train_cat_ohe, y_train)
array([0.53642384, 0.53642384, 0.50993377, 0.51333333, 0.47333333])
Are we breaking the golden rule here? Let’s do this properly with a pipeline.
# Code to create a pipeline for OHE and KNN
pipe_ohe_knn = make_pipeline(OneHotEncoder(), KNeighborsClassifier())
cross_val_score(pipe_ohe_knn, X_train_cat, y_train)
/Users/kvarada/miniconda3/envs/cpsc330/lib/python3.10/site-packages/sklearn/model_selection/_validation.py:839: UserWarning: Scoring failed. The score on this train-test partition for these parameters will be set to nan. Details:
Traceback (most recent call last):
File "/Users/kvarada/miniconda3/envs/cpsc330/lib/python3.10/site-packages/sklearn/metrics/_scorer.py", line 140, in __call__
score = scorer(estimator, *args, **routed_params.get(name).score)
File "/Users/kvarada/miniconda3/envs/cpsc330/lib/python3.10/site-packages/sklearn/metrics/_scorer.py", line 527, in __call__
return estimator.score(*args, **kwargs)
File "/Users/kvarada/miniconda3/envs/cpsc330/lib/python3.10/site-packages/sklearn/pipeline.py", line 756, in score
Xt = transform.transform(Xt)
File "/Users/kvarada/miniconda3/envs/cpsc330/lib/python3.10/site-packages/sklearn/utils/_set_output.py", line 157, in wrapped
data_to_wrap = f(self, X, *args, **kwargs)
File "/Users/kvarada/miniconda3/envs/cpsc330/lib/python3.10/site-packages/sklearn/preprocessing/_encoders.py", line 1027, in transform
X_int, X_mask = self._transform(
File "/Users/kvarada/miniconda3/envs/cpsc330/lib/python3.10/site-packages/sklearn/preprocessing/_encoders.py", line 200, in _transform
raise ValueError(msg)
ValueError: Found unknown categories ['Quebecois'] in column 1 during transform
warnings.warn(
array([ nan, 0.53642384, 0.56291391, 0.54666667, 0.56 ])
What’s wrong here?
How can we fix this?
# Fix the OHE
pipe_ohe_knn = make_pipeline(OneHotEncoder(handle_unknown="ignore"), KNeighborsClassifier())
cross_val_score(pipe_ohe_knn, X_train_cat, y_train).mean()
0.5537836644591612
Right now we are working with numeric and categorical features separately. But ideally when we create a model, we need to use all these features together.
Enter column transformer!
How can we vertically stack
preprocessed numeric features
preprocessed binary features, and
preprocessed categorical features?
Let’s define a column transformer.
from sklearn.compose import make_column_transformer
numeric_transformer = make_pipeline(SimpleImputer(strategy="median"), StandardScaler())
binary_transformer = make_pipeline(SimpleImputer(strategy="most_frequent"), OneHotEncoder(drop="if_binary"))
categorical_transformer = make_pipeline(SimpleImputer(strategy="most_frequent"), OneHotEncoder(handle_unknown="ignore", sparse_output=False))
preprocessor = make_column_transformer(
(numeric_transformer, numeric_feats),
(binary_transformer, binary_feats),
(categorical_transformer, categorical_feats),
)
How does the transformed data look like?
transformed = preprocessor.fit_transform(X_train)
preprocessor
ColumnTransformer(transformers=[('pipeline-1',
Pipeline(steps=[('simpleimputer',
SimpleImputer(strategy='median')),
('standardscaler',
StandardScaler())]),
['age', 'n_people', 'price']),
('pipeline-2',
Pipeline(steps=[('simpleimputer',
SimpleImputer(strategy='most_frequent')),
('onehotencoder',
OneHotEncoder(drop='if_binary'))]),
['good_server']),
('pipeline-3',
Pipeline(steps=[('simpleimputer',
SimpleImputer(strategy='most_frequent')),
('onehotencoder',
OneHotEncoder(handle_unknown='ignore',
sparse_output=False))]),
['north_america', 'food_type'])])In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook. On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
ColumnTransformer(transformers=[('pipeline-1',
Pipeline(steps=[('simpleimputer',
SimpleImputer(strategy='median')),
('standardscaler',
StandardScaler())]),
['age', 'n_people', 'price']),
('pipeline-2',
Pipeline(steps=[('simpleimputer',
SimpleImputer(strategy='most_frequent')),
('onehotencoder',
OneHotEncoder(drop='if_binary'))]),
['good_server']),
('pipeline-3',
Pipeline(steps=[('simpleimputer',
SimpleImputer(strategy='most_frequent')),
('onehotencoder',
OneHotEncoder(handle_unknown='ignore',
sparse_output=False))]),
['north_america', 'food_type'])])['age', 'n_people', 'price']
SimpleImputer(strategy='median')
StandardScaler()
['good_server']
SimpleImputer(strategy='most_frequent')
OneHotEncoder(drop='if_binary')
['north_america', 'food_type']
SimpleImputer(strategy='most_frequent')
OneHotEncoder(handle_unknown='ignore', sparse_output=False)
# Getting feature names from a column transformer
ohe_feat_names = preprocessor.named_transformers_['pipeline-3']['onehotencoder'].get_feature_names_out(categorical_feats).tolist()
ohe_feat_names
["north_america_Don't want to share",
'north_america_No',
'north_america_Yes',
'food_type_Canadian/American',
'food_type_Chinese',
'food_type_Fusion',
'food_type_Indian',
'food_type_Italian',
'food_type_Mexican',
'food_type_Other',
'food_type_Quebecois',
'food_type_Thai']
# feat_names = numeric_feats + binary_feats + ohe_feat_names
# pd.DataFrame(preprocessor.fit_transform(X_train), columns = feat_names)
We have new columns for the categorical features. Let’s create a pipeline with the preprocessor and SVC.
svc_num_cat_pipe = make_pipeline(preprocessor, SVC())
cross_val_score(svc_num_cat_pipe, X_train, y_train).mean()
0.6825960264900662
We are getting better results!
Incorporating text features#
We haven’t incorporated the comments feature into our pipeline yet, even though it holds significant value in indicating whether the restaurant was liked or not.
X_train
| north_america | eat_out_freq | age | n_people | price | food_type | noise_level | good_server | comments | restaurant_name | |
|---|---|---|---|---|---|---|---|---|---|---|
| 80 | No | 2.0 | 21 | 30.0 | 2200.0 | Chinese | high | No | The environment was very not clean. The food tasted awful. | NaN |
| 934 | Yes | 4.0 | 21 | 30.0 | 3000.0 | Canadian/American | low | Yes | The building and the room gave a very comfy feeling. Immediately after sitting down it felt like we were right at home. | NaN |
| 911 | No | 4.0 | 20 | 40.0 | 2500.0 | Canadian/American | medium | Yes | I was hungry | Chambar |
| 459 | Yes | 5.0 | 21 | NaN | NaN | Quebecois | NaN | NaN | NaN | NaN |
| 62 | Yes | 2.0 | 24 | 20.0 | 3000.0 | Indian | high | Yes | bad taste | east is east |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 106 | No | 3.0 | 27 | 10.0 | 1500.0 | Chinese | medium | Yes | Food wasn't great. | NaN |
| 333 | No | 1.0 | 24 | 12.0 | 800.0 | Other | medium | Yes | NaN | NaN |
| 393 | Yes | 4.0 | 20 | 5.0 | 1500.0 | Canadian/American | low | No | NaN | NaN |
| 376 | Yes | 5.0 | 20 | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 525 | Don't want to share | 4.0 | 20 | 50.0 | 3000.0 | Chinese | high | Yes | NaN | Haidilao |
753 rows × 10 columns
Let’s create bag-of-words representation of the comments feature. But first we need to impute the rows where there are no comments. There is a small complication if we want to put SimpleImputer and CountVectorizer in a pipeline.
SimpleImputertakes a 2D array as input and produced 2D array as output.CountVectorizertakes a 1D array as input.
To deal with this, we will use sklearn’s FunctionTransformer to convert the 2D output of SimpleImputer into a 1D array which can be passed to CountVectorizer as input.
from sklearn.preprocessing import FunctionTransformer
from sklearn.feature_extraction.text import CountVectorizer
reshape_for_countvectorizer = FunctionTransformer(lambda X: X.squeeze(), validate=False)
text_transformer = make_pipeline(SimpleImputer(strategy="constant", fill_value="missing"),
reshape_for_countvectorizer,
CountVectorizer(max_features=100, stop_words="english"))
text_pipe = make_pipeline(text_transformer, SVC())
cross_val_score(text_pipe, X_train[['comments']], y_train).mean()
0.6467019867549668
Pretty good scores just with text features! Do we get better scores if we combine all features? Let’s define a column transformer which carries out
imputation and scaling on numeric features
imputation and one-hot encoding with
drop="if_binary"on binary featuresimputation and one-hot encoding with
handle_unknown="ignore"on categorical featuresimputation, reshaping, and bag-of-words transformation on the text feature
from sklearn.feature_extraction.text import CountVectorizer
text_feat = ['comments']
preprocessor = make_column_transformer(
(numeric_transformer, numeric_feats),
(binary_transformer, binary_feats),
(categorical_transformer, categorical_feats),
(text_transformer, text_feat)
)
preprocessor.fit_transform(X_train)
<753x116 sparse matrix of type '<class 'numpy.float64'>'
with 5670 stored elements in Compressed Sparse Row format>
svc_num_cat_text_pipe = make_pipeline(preprocessor, SVC())
cross_val_score(svc_num_cat_text_pipe, X_train, y_train).mean()
0.6972185430463577
No big improvement when we combine all features.
