
Lecture 5: Preprocessing and sklearn pipelines#
UBC 2024-25
Imports, LOs#
Imports#
import os
import sys
import time
import matplotlib.pyplot as plt
%matplotlib inline
import numpy as np
import pandas as pd
from IPython.display import HTML
sys.path.append(os.path.join(os.path.abspath(".."), "code"))
import mglearn
from IPython.display import display
from plotting_functions import *
# Classifiers and regressors
from sklearn.dummy import DummyClassifier, DummyRegressor
# Preprocessing and pipeline
from sklearn.impute import SimpleImputer
# train test split and cross validation
from sklearn.model_selection import cross_val_score, cross_validate, train_test_split
from sklearn.neighbors import KNeighborsClassifier, KNeighborsRegressor
from sklearn.pipeline import Pipeline
from sklearn.preprocessing import (
MinMaxScaler,
OneHotEncoder,
OrdinalEncoder,
StandardScaler,
)
from sklearn.svm import SVC
from sklearn.tree import DecisionTreeClassifier
from utils import *
%matplotlib inline
pd.set_option("display.max_colwidth", 200)
DATA_DIR = "../data/"
Learning outcomes#
From this lecture, you will be able to
explain motivation for preprocessing in supervised machine learning;
identify when to implement feature transformations such as imputation, scaling, and one-hot encoding in a machine learning model development pipeline;
use
sklearntransformers for applying feature transformations on your dataset;discuss golden rule in the context of feature transformations;
use
sklearn.pipeline.Pipelineandsklearn.pipeline.make_pipelineto build a preliminary machine learning pipeline.
❓❓ Questions for you#
(iClicker) Exercise 5.1#
iClicker cloud join link: https://join.iclicker.com/VYFJ
Take a guess: In your machine learning project, how much time will you typically spend on data preparation and transformation?
(A) ~80% of the project time
(B) ~20% of the project time
(C) ~50% of the project time
(D) None. Most of the time will be spent on model building
The question is adapted from here.
Motivation and big picture [video]#
So far we have seen
Three ML models (decision trees, \(k\)-NNs, SVMs with RBF kernel)
ML fundamentals (train-validation-test split, cross-validation, the fundamental tradeoff, the golden rule)
Are we ready to do machine learning on real-world datasets?
Very often real-world datasets need preprocessing before we use them to build ML models.
Example: \(k\)-nearest neighbours on the Spotify dataset#
In HW2 you used
DecisionTreeClassifierto predict whether the user would like a particular song or not.Can we use \(k\)-NN classifier for this task?
Intuition: To predict whether the user likes a particular song or not (query point)
find the songs that are closest to the query point
let them vote on the target
take the majority vote as the target for the query point
In order to run the code below, you need to download the dataset from Kaggle.
spotify_df = pd.read_csv(DATA_DIR + "spotify.csv", index_col=0)
train_df, test_df = train_test_split(spotify_df, test_size=0.20, random_state=123)
X_train, y_train = (
train_df.drop(columns=["song_title", "artist", "target"]),
train_df["target"],
)
X_test, y_test = (
test_df.drop(columns=["song_title", "artist", "target"]),
test_df["target"],
)
dummy = DummyClassifier(strategy="most_frequent")
scores = cross_validate(dummy, X_train, y_train, return_train_score=True)
print("Mean validation score %0.3f" % (np.mean(scores["test_score"])))
pd.DataFrame(scores)
Mean validation score 0.508
| fit_time | score_time | test_score | train_score | |
|---|---|---|---|---|
| 0 | 0.001772 | 0.001699 | 0.507740 | 0.507752 |
| 1 | 0.000935 | 0.000662 | 0.507740 | 0.507752 |
| 2 | 0.000815 | 0.000617 | 0.507740 | 0.507752 |
| 3 | 0.000963 | 0.000847 | 0.506211 | 0.508133 |
| 4 | 0.001010 | 0.001963 | 0.509317 | 0.507359 |
knn = KNeighborsClassifier()
scores = cross_validate(knn, X_train, y_train, return_train_score=True)
print("Mean validation score %0.3f" % (np.mean(scores["test_score"])))
pd.DataFrame(scores)
Mean validation score 0.546
| fit_time | score_time | test_score | train_score | |
|---|---|---|---|---|
| 0 | 0.024021 | 0.025336 | 0.563467 | 0.717829 |
| 1 | 0.002007 | 0.013260 | 0.535604 | 0.721705 |
| 2 | 0.001785 | 0.013105 | 0.529412 | 0.708527 |
| 3 | 0.001765 | 0.012947 | 0.537267 | 0.721921 |
| 4 | 0.001753 | 0.013504 | 0.562112 | 0.711077 |
two_songs = X_train.sample(2, random_state=42)
two_songs
| acousticness | danceability | duration_ms | energy | instrumentalness | key | liveness | loudness | mode | speechiness | tempo | time_signature | valence | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 842 | 0.229000 | 0.494 | 147893 | 0.666 | 0.000057 | 9 | 0.0469 | -9.743 | 0 | 0.0351 | 140.832 | 4.0 | 0.704 |
| 654 | 0.000289 | 0.771 | 227143 | 0.949 | 0.602000 | 8 | 0.5950 | -4.712 | 1 | 0.1750 | 111.959 | 4.0 | 0.372 |
euclidean_distances(two_songs)
Intel MKL WARNING: Support of Intel(R) Streaming SIMD Extensions 4.2 (Intel(R) SSE4.2) enabled only processors has been deprecated. Intel oneAPI Math Kernel Library 2025.0 will require Intel(R) Advanced Vector Extensions (Intel(R) AVX) instructions.
array([[ 0. , 79250.00543825],
[79250.00543825, 0. ]])
Let’s consider only two features: duration_ms and tempo.
two_songs_subset = two_songs[["duration_ms", "tempo"]]
two_songs_subset
| duration_ms | tempo | |
|---|---|---|
| 842 | 147893 | 140.832 |
| 654 | 227143 | 111.959 |
euclidean_distances(two_songs_subset)
Intel MKL WARNING: Support of Intel(R) Streaming SIMD Extensions 4.2 (Intel(R) SSE4.2) enabled only processors has been deprecated. Intel oneAPI Math Kernel Library 2025.0 will require Intel(R) Advanced Vector Extensions (Intel(R) AVX) instructions.
array([[ 0. , 79250.00525962],
[79250.00525962, 0. ]])
Do you see any problem?
The distance is completely dominated by the the features with larger values
The features with smaller values are being ignored.
Does it matter?
Yes! Scale is based on how data was collected.
Features on a smaller scale can be highly informative and there is no good reason to ignore them.
We want our model to be robust and not sensitive to the scale.
Was this a problem for decision trees?
Scaling using scikit-learn’s StandardScaler#
We’ll use
scikit-learn’sStandardScaler, which is atransformer.Only focus on the syntax for now. We’ll talk about scaling in a bit.
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler() # create feature trasformer object
scaler.fit(X_train) # fitting the transformer on the train split
X_train_scaled = scaler.transform(X_train) # transforming the train split
X_test_scaled = scaler.transform(X_test) # transforming the test split
X_train # original X_train
| acousticness | danceability | duration_ms | energy | instrumentalness | key | liveness | loudness | mode | speechiness | tempo | time_signature | valence | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1505 | 0.004770 | 0.585 | 214740 | 0.614 | 0.000155 | 10 | 0.0762 | -5.594 | 0 | 0.0370 | 114.059 | 4.0 | 0.2730 |
| 813 | 0.114000 | 0.665 | 216728 | 0.513 | 0.303000 | 0 | 0.1220 | -7.314 | 1 | 0.3310 | 100.344 | 3.0 | 0.0373 |
| 615 | 0.030200 | 0.798 | 216585 | 0.481 | 0.000000 | 7 | 0.1280 | -10.488 | 1 | 0.3140 | 127.136 | 4.0 | 0.6400 |
| 319 | 0.106000 | 0.912 | 194040 | 0.317 | 0.000208 | 6 | 0.0723 | -12.719 | 0 | 0.0378 | 99.346 | 4.0 | 0.9490 |
| 320 | 0.021100 | 0.697 | 236456 | 0.905 | 0.893000 | 6 | 0.1190 | -7.787 | 0 | 0.0339 | 119.977 | 4.0 | 0.3110 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 2012 | 0.001060 | 0.584 | 274404 | 0.932 | 0.002690 | 1 | 0.1290 | -3.501 | 1 | 0.3330 | 74.976 | 4.0 | 0.2110 |
| 1346 | 0.000021 | 0.535 | 203500 | 0.974 | 0.000149 | 10 | 0.2630 | -3.566 | 0 | 0.1720 | 116.956 | 4.0 | 0.4310 |
| 1406 | 0.503000 | 0.410 | 256333 | 0.648 | 0.000000 | 7 | 0.2190 | -4.469 | 1 | 0.0362 | 60.391 | 4.0 | 0.3420 |
| 1389 | 0.705000 | 0.894 | 222307 | 0.161 | 0.003300 | 4 | 0.3120 | -14.311 | 1 | 0.0880 | 104.968 | 4.0 | 0.8180 |
| 1534 | 0.623000 | 0.470 | 394920 | 0.156 | 0.187000 | 2 | 0.1040 | -17.036 | 1 | 0.0399 | 118.176 | 4.0 | 0.0591 |
1613 rows × 13 columns
Let’s examine transformed value of the energy feature in the first row.
X_train["energy"].iloc[0]
0.614
(X_train["energy"].iloc[0] - np.mean(X_train["energy"])) / X_train["energy"].std()
-0.3180174485124284
pd.DataFrame(X_train_scaled, columns=X_train.columns, index=X_train.index).head().round(
3
)
| acousticness | danceability | duration_ms | energy | instrumentalness | key | liveness | loudness | mode | speechiness | tempo | time_signature | valence | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1505 | -0.698 | -0.195 | -0.399 | -0.318 | -0.492 | 1.276 | -0.738 | 0.396 | -1.281 | -0.618 | -0.294 | 0.139 | -0.908 |
| 813 | -0.276 | 0.296 | -0.374 | -0.796 | 0.598 | -1.487 | -0.439 | -0.052 | 0.781 | 2.728 | -0.803 | -3.781 | -1.861 |
| 615 | -0.600 | 1.111 | -0.376 | -0.947 | -0.493 | 0.447 | -0.400 | -0.879 | 0.781 | 2.535 | 0.191 | 0.139 | 0.576 |
| 319 | -0.307 | 1.809 | -0.654 | -1.722 | -0.492 | 0.170 | -0.763 | -1.461 | -1.281 | -0.609 | -0.840 | 0.139 | 1.825 |
| 320 | -0.635 | 0.492 | -0.131 | 1.057 | 2.723 | 0.170 | -0.458 | -0.176 | -1.281 | -0.653 | -0.074 | 0.139 | -0.754 |
fit and transform paradigm for transformers#
sklearnusesfitandtransformparadigms for feature transformations.We
fitthe transformer on the train split and then transform the train split as well as the test split.We apply the same transformations on the test split.
sklearn API summary: estimators#
Suppose model is a classification or regression model.
model.fit(X_train, y_train)
X_train_predictions = model.predict(X_train)
X_test_predictions = model.predict(X_test)
sklearn API summary: transformers#
Suppose transformer is a transformer used to change the input representation, for example, to tackle missing values or to scales numeric features.
transformer.fit(X_train, [y_train])
X_train_transformed = transformer.transform(X_train)
X_test_transformed = transformer.transform(X_test)
You can pass
y_traininfitbut it’s usually ignored. It allows you to pass it just to be consistent with usual usage ofsklearn’sfitmethod.You can also carry out fitting and transforming in one call using
fit_transform. But be mindful to use it only on the train split and not on the test split.
Do you expect
DummyClassifierresults to change after scaling the data?Let’s check whether scaling makes any difference for \(k\)-NNs.
knn_unscaled = KNeighborsClassifier()
knn_unscaled.fit(X_train, y_train)
print("Train score: %0.3f" % (knn_unscaled.score(X_train, y_train)))
print("Test score: %0.3f" % (knn_unscaled.score(X_test, y_test)))
Train score: 0.726
Test score: 0.552
knn_scaled = KNeighborsClassifier()
knn_scaled.fit(X_train_scaled, y_train)
print("Train score: %0.3f" % (knn_scaled.score(X_train_scaled, y_train)))
print("Test score: %0.3f" % (knn_scaled.score(X_test_scaled, y_test)))
Train score: 0.798
Test score: 0.686
The scores with scaled data are better compared to the unscaled data in case of \(k\)-NNs.
I am not carrying out cross-validation here for a reason that we’ll look into soon.
Note that I am a bit sloppy here and using the test set several times for teaching purposes. But when you build an ML pipeline, please do assessment on the test set only once.
Common preprocessing techniques#
Some commonly performed feature transformation include:
Imputation: Tackling missing values
Scaling: Scaling of numeric features
One-hot encoding: Tackling categorical variables
We can have one lecture on each of them! In this lesson our goal is to getting familiar with them so that we can use them to build ML pipelines.
In the next part of this lecture, we’ll build an ML pipeline using California housing prices regression dataset. In the process, we will talk about different feature transformations and how can we apply them so that we do not violate the golden rule.
Imputation and scaling [video]#
Dataset, splitting, and baseline#
We’ll be working on California housing prices regression dataset to demonstrate these feature transformation techniques. The task is to predict median house values in Californian districts, given a number of features from these districts. If you are running the notebook on your own, you’ll have to download the data and put it in the data directory.
housing_df = pd.read_csv(DATA_DIR + "housing.csv")
train_df, test_df = train_test_split(housing_df, test_size=0.1, random_state=123)
train_df.head()
| longitude | latitude | housing_median_age | total_rooms | total_bedrooms | population | households | median_income | median_house_value | ocean_proximity | |
|---|---|---|---|---|---|---|---|---|---|---|
| 6051 | -117.75 | 34.04 | 22.0 | 2948.0 | 636.0 | 2600.0 | 602.0 | 3.1250 | 113600.0 | INLAND |
| 20113 | -119.57 | 37.94 | 17.0 | 346.0 | 130.0 | 51.0 | 20.0 | 3.4861 | 137500.0 | INLAND |
| 14289 | -117.13 | 32.74 | 46.0 | 3355.0 | 768.0 | 1457.0 | 708.0 | 2.6604 | 170100.0 | NEAR OCEAN |
| 13665 | -117.31 | 34.02 | 18.0 | 1634.0 | 274.0 | 899.0 | 285.0 | 5.2139 | 129300.0 | INLAND |
| 14471 | -117.23 | 32.88 | 18.0 | 5566.0 | 1465.0 | 6303.0 | 1458.0 | 1.8580 | 205000.0 | NEAR OCEAN |
Let’s add some new features to the dataset which could help predicting the target: median_house_value.
train_df = train_df.assign(
rooms_per_household=train_df["total_rooms"] / train_df["households"]
)
test_df = test_df.assign(
rooms_per_household=test_df["total_rooms"] / test_df["households"]
)
train_df = train_df.assign(
bedrooms_per_household=train_df["total_bedrooms"] / train_df["households"]
)
test_df = test_df.assign(
bedrooms_per_household=test_df["total_bedrooms"] / test_df["households"]
)
train_df = train_df.assign(
population_per_household=train_df["population"] / train_df["households"]
)
test_df = test_df.assign(
population_per_household=test_df["population"] / test_df["households"]
)
train_df.head()
| longitude | latitude | housing_median_age | total_rooms | total_bedrooms | population | households | median_income | median_house_value | ocean_proximity | rooms_per_household | bedrooms_per_household | population_per_household | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 6051 | -117.75 | 34.04 | 22.0 | 2948.0 | 636.0 | 2600.0 | 602.0 | 3.1250 | 113600.0 | INLAND | 4.897010 | 1.056478 | 4.318937 |
| 20113 | -119.57 | 37.94 | 17.0 | 346.0 | 130.0 | 51.0 | 20.0 | 3.4861 | 137500.0 | INLAND | 17.300000 | 6.500000 | 2.550000 |
| 14289 | -117.13 | 32.74 | 46.0 | 3355.0 | 768.0 | 1457.0 | 708.0 | 2.6604 | 170100.0 | NEAR OCEAN | 4.738701 | 1.084746 | 2.057910 |
| 13665 | -117.31 | 34.02 | 18.0 | 1634.0 | 274.0 | 899.0 | 285.0 | 5.2139 | 129300.0 | INLAND | 5.733333 | 0.961404 | 3.154386 |
| 14471 | -117.23 | 32.88 | 18.0 | 5566.0 | 1465.0 | 6303.0 | 1458.0 | 1.8580 | 205000.0 | NEAR OCEAN | 3.817558 | 1.004801 | 4.323045 |
train_df = train_df.drop(columns=["population", "total_rooms", "total_bedrooms"])
test_df = test_df.drop(columns=["population", "total_rooms", "total_bedrooms"])
When is it OK to do things before splitting?#
Here it would have been OK to add new features before splitting because we are not using any global information in the data but only looking at one row at a time.
But just to be safe and to avoid accidentally breaking the golden rule, it’s better to do it after splitting.
Why are we removing
total_rooms,total_bedrooms, andpopulationcolumns?That information is available in the new features, and it is desirable to remove unnecessary features, especially when correlated with others.
EDA#
train_df.head()
| longitude | latitude | housing_median_age | households | median_income | median_house_value | ocean_proximity | rooms_per_household | bedrooms_per_household | population_per_household | |
|---|---|---|---|---|---|---|---|---|---|---|
| 6051 | -117.75 | 34.04 | 22.0 | 602.0 | 3.1250 | 113600.0 | INLAND | 4.897010 | 1.056478 | 4.318937 |
| 20113 | -119.57 | 37.94 | 17.0 | 20.0 | 3.4861 | 137500.0 | INLAND | 17.300000 | 6.500000 | 2.550000 |
| 14289 | -117.13 | 32.74 | 46.0 | 708.0 | 2.6604 | 170100.0 | NEAR OCEAN | 4.738701 | 1.084746 | 2.057910 |
| 13665 | -117.31 | 34.02 | 18.0 | 285.0 | 5.2139 | 129300.0 | INLAND | 5.733333 | 0.961404 | 3.154386 |
| 14471 | -117.23 | 32.88 | 18.0 | 1458.0 | 1.8580 | 205000.0 | NEAR OCEAN | 3.817558 | 1.004801 | 4.323045 |
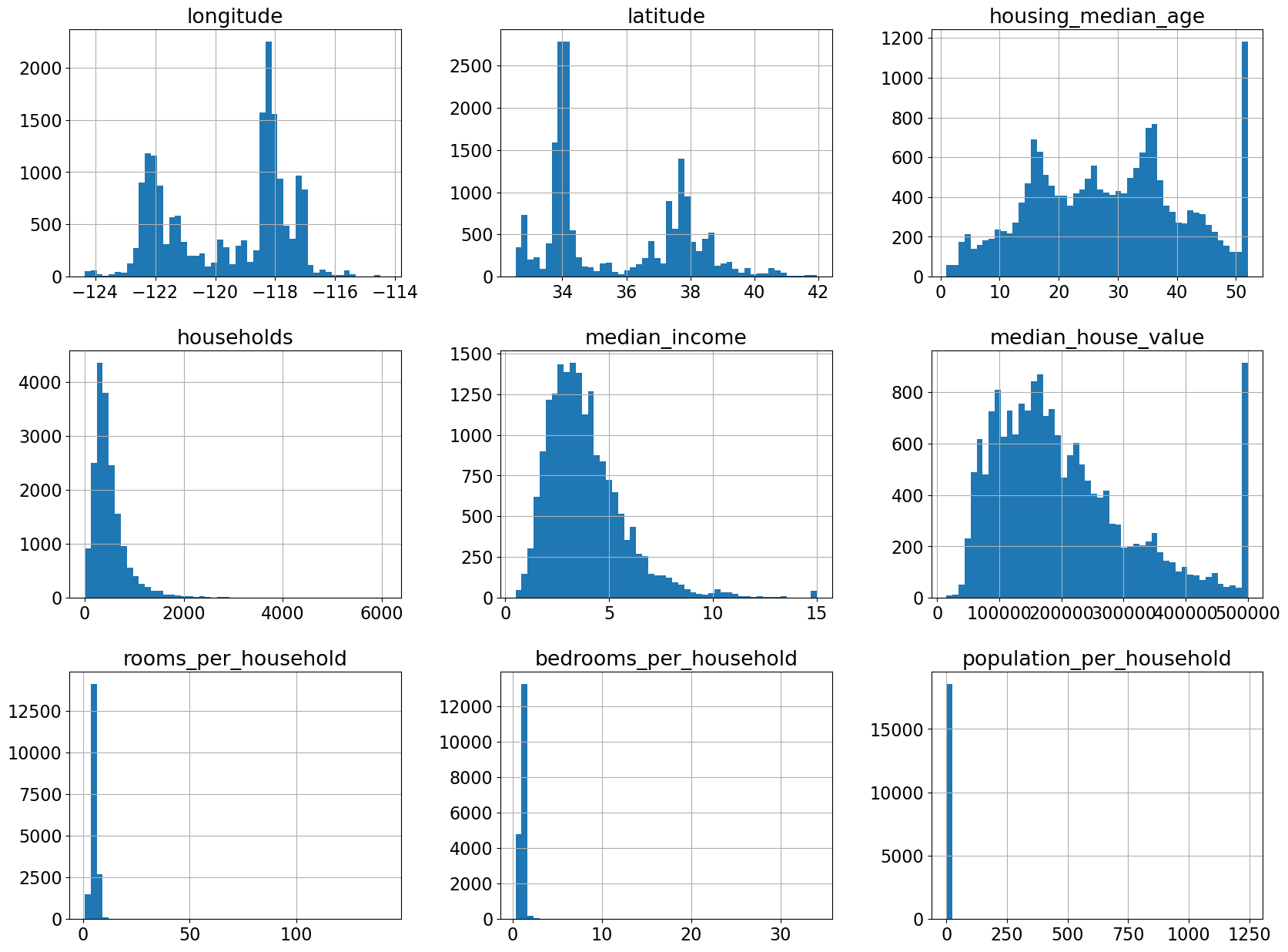
The feature scales are quite different.
train_df.info()
<class 'pandas.core.frame.DataFrame'>
Index: 18576 entries, 6051 to 19966
Data columns (total 10 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 longitude 18576 non-null float64
1 latitude 18576 non-null float64
2 housing_median_age 18576 non-null float64
3 households 18576 non-null float64
4 median_income 18576 non-null float64
5 median_house_value 18576 non-null float64
6 ocean_proximity 18576 non-null object
7 rooms_per_household 18576 non-null float64
8 bedrooms_per_household 18391 non-null float64
9 population_per_household 18576 non-null float64
dtypes: float64(9), object(1)
memory usage: 1.6+ MB
We have one categorical feature and all other features are numeric features.
train_df.describe()
| longitude | latitude | housing_median_age | households | median_income | median_house_value | rooms_per_household | bedrooms_per_household | population_per_household | |
|---|---|---|---|---|---|---|---|---|---|
| count | 18576.000000 | 18576.000000 | 18576.000000 | 18576.000000 | 18576.000000 | 18576.000000 | 18576.000000 | 18391.000000 | 18576.000000 |
| mean | -119.565888 | 35.627966 | 28.622255 | 500.061100 | 3.862552 | 206292.067991 | 5.426067 | 1.097516 | 3.052349 |
| std | 1.999622 | 2.134658 | 12.588307 | 383.044313 | 1.892491 | 115083.856175 | 2.512319 | 0.486266 | 10.020873 |
| min | -124.350000 | 32.540000 | 1.000000 | 1.000000 | 0.499900 | 14999.000000 | 0.846154 | 0.333333 | 0.692308 |
| 25% | -121.790000 | 33.930000 | 18.000000 | 280.000000 | 2.560225 | 119400.000000 | 4.439360 | 1.005888 | 2.430323 |
| 50% | -118.490000 | 34.250000 | 29.000000 | 410.000000 | 3.527500 | 179300.000000 | 5.226415 | 1.048860 | 2.818868 |
| 75% | -118.010000 | 37.710000 | 37.000000 | 606.000000 | 4.736900 | 263600.000000 | 6.051620 | 1.099723 | 3.283921 |
| max | -114.310000 | 41.950000 | 52.000000 | 6082.000000 | 15.000100 | 500001.000000 | 141.909091 | 34.066667 | 1243.333333 |
Seems like bedrooms_per_household column has some missing values.
This must have affected our new feature
bedrooms_per_householdas well.
housing_df["total_bedrooms"].isnull().sum()
207
## (optional)
train_df.hist(bins=50, figsize=(20, 15));
What transformations we need to apply on the dataset?#
Here is what we see from the EDA.
Some missing values in
total_bedroomscolumnScales are quite different across columns.
Categorical variable
ocean_proximity
Read about preprocessing techniques implemented in scikit-learn.
# We are droping the categorical variable ocean_proximity for now. We'll come back to it in a bit.
X_train = train_df.drop(columns=["median_house_value", "ocean_proximity"])
y_train = train_df["median_house_value"]
X_test = test_df.drop(columns=["median_house_value", "ocean_proximity"])
y_test = test_df["median_house_value"]
Let’s first run our baseline model DummyRegressor#
results_dict = {} # dictionary to store our results for different models
def mean_std_cross_val_scores(model, X_train, y_train, **kwargs):
"""
Returns mean and std of cross validation
Parameters
----------
model :
scikit-learn model
X_train : numpy array or pandas DataFrame
X in the training data
y_train :
y in the training data
Returns
----------
pandas Series with mean scores from cross_validation
"""
scores = cross_validate(model, X_train, y_train, **kwargs)
mean_scores = pd.DataFrame(scores).mean()
std_scores = pd.DataFrame(scores).std()
out_col = []
for i in range(len(mean_scores)):
out_col.append((f"%0.3f (+/- %0.3f)" % (mean_scores[i], std_scores[i])))
return pd.Series(data=out_col, index=mean_scores.index)
dummy = DummyRegressor(strategy="median")
results_dict["dummy"] = mean_std_cross_val_scores(
dummy, X_train, y_train, return_train_score=True
)
/var/folders/7l/2m7m0lw97rvf654x1cwtdfmr0000gr/T/ipykernel_53933/4158382658.py:26: FutureWarning: Series.__getitem__ treating keys as positions is deprecated. In a future version, integer keys will always be treated as labels (consistent with DataFrame behavior). To access a value by position, use `ser.iloc[pos]`
out_col.append((f"%0.3f (+/- %0.3f)" % (mean_scores[i], std_scores[i])))
pd.DataFrame(results_dict)
| dummy | |
|---|---|
| fit_time | 0.003 (+/- 0.003) |
| score_time | 0.004 (+/- 0.006) |
| test_score | -0.055 (+/- 0.012) |
| train_score | -0.055 (+/- 0.001) |
Imputation#
X_train
| longitude | latitude | housing_median_age | households | median_income | rooms_per_household | bedrooms_per_household | population_per_household | |
|---|---|---|---|---|---|---|---|---|
| 6051 | -117.75 | 34.04 | 22.0 | 602.0 | 3.1250 | 4.897010 | 1.056478 | 4.318937 |
| 20113 | -119.57 | 37.94 | 17.0 | 20.0 | 3.4861 | 17.300000 | 6.500000 | 2.550000 |
| 14289 | -117.13 | 32.74 | 46.0 | 708.0 | 2.6604 | 4.738701 | 1.084746 | 2.057910 |
| 13665 | -117.31 | 34.02 | 18.0 | 285.0 | 5.2139 | 5.733333 | 0.961404 | 3.154386 |
| 14471 | -117.23 | 32.88 | 18.0 | 1458.0 | 1.8580 | 3.817558 | 1.004801 | 4.323045 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 7763 | -118.10 | 33.91 | 36.0 | 130.0 | 3.6389 | 5.584615 | NaN | 3.769231 |
| 15377 | -117.24 | 33.37 | 14.0 | 779.0 | 4.5391 | 6.016688 | 1.017972 | 3.127086 |
| 17730 | -121.76 | 37.33 | 5.0 | 697.0 | 5.6306 | 5.958393 | 1.031564 | 3.493544 |
| 15725 | -122.44 | 37.78 | 44.0 | 326.0 | 3.8750 | 4.739264 | 1.024540 | 1.720859 |
| 19966 | -119.08 | 36.21 | 20.0 | 348.0 | 2.5156 | 5.491379 | 1.117816 | 3.566092 |
18576 rows × 8 columns
knn = KNeighborsRegressor()
knn.fit(X_train, y_train)
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
Cell In[34], line 2
1 knn = KNeighborsRegressor()
----> 2 knn.fit(X_train, y_train)
File ~/opt/anaconda3/envs/cpsc330-24/lib/python3.12/site-packages/sklearn/base.py:1473, in _fit_context.<locals>.decorator.<locals>.wrapper(estimator, *args, **kwargs)
1466 estimator._validate_params()
1468 with config_context(
1469 skip_parameter_validation=(
1470 prefer_skip_nested_validation or global_skip_validation
1471 )
1472 ):
-> 1473 return fit_method(estimator, *args, **kwargs)
File ~/opt/anaconda3/envs/cpsc330-24/lib/python3.12/site-packages/sklearn/neighbors/_regression.py:223, in KNeighborsRegressor.fit(self, X, y)
201 @_fit_context(
202 # KNeighborsRegressor.metric is not validated yet
203 prefer_skip_nested_validation=False
204 )
205 def fit(self, X, y):
206 """Fit the k-nearest neighbors regressor from the training dataset.
207
208 Parameters
(...)
221 The fitted k-nearest neighbors regressor.
222 """
--> 223 return self._fit(X, y)
File ~/opt/anaconda3/envs/cpsc330-24/lib/python3.12/site-packages/sklearn/neighbors/_base.py:475, in NeighborsBase._fit(self, X, y)
473 if self._get_tags()["requires_y"]:
474 if not isinstance(X, (KDTree, BallTree, NeighborsBase)):
--> 475 X, y = self._validate_data(
476 X, y, accept_sparse="csr", multi_output=True, order="C"
477 )
479 if is_classifier(self):
480 # Classification targets require a specific format
481 if y.ndim == 1 or y.ndim == 2 and y.shape[1] == 1:
File ~/opt/anaconda3/envs/cpsc330-24/lib/python3.12/site-packages/sklearn/base.py:650, in BaseEstimator._validate_data(self, X, y, reset, validate_separately, cast_to_ndarray, **check_params)
648 y = check_array(y, input_name="y", **check_y_params)
649 else:
--> 650 X, y = check_X_y(X, y, **check_params)
651 out = X, y
653 if not no_val_X and check_params.get("ensure_2d", True):
File ~/opt/anaconda3/envs/cpsc330-24/lib/python3.12/site-packages/sklearn/utils/validation.py:1301, in check_X_y(X, y, accept_sparse, accept_large_sparse, dtype, order, copy, force_writeable, force_all_finite, ensure_2d, allow_nd, multi_output, ensure_min_samples, ensure_min_features, y_numeric, estimator)
1296 estimator_name = _check_estimator_name(estimator)
1297 raise ValueError(
1298 f"{estimator_name} requires y to be passed, but the target y is None"
1299 )
-> 1301 X = check_array(
1302 X,
1303 accept_sparse=accept_sparse,
1304 accept_large_sparse=accept_large_sparse,
1305 dtype=dtype,
1306 order=order,
1307 copy=copy,
1308 force_writeable=force_writeable,
1309 force_all_finite=force_all_finite,
1310 ensure_2d=ensure_2d,
1311 allow_nd=allow_nd,
1312 ensure_min_samples=ensure_min_samples,
1313 ensure_min_features=ensure_min_features,
1314 estimator=estimator,
1315 input_name="X",
1316 )
1318 y = _check_y(y, multi_output=multi_output, y_numeric=y_numeric, estimator=estimator)
1320 check_consistent_length(X, y)
File ~/opt/anaconda3/envs/cpsc330-24/lib/python3.12/site-packages/sklearn/utils/validation.py:1064, in check_array(array, accept_sparse, accept_large_sparse, dtype, order, copy, force_writeable, force_all_finite, ensure_2d, allow_nd, ensure_min_samples, ensure_min_features, estimator, input_name)
1058 raise ValueError(
1059 "Found array with dim %d. %s expected <= 2."
1060 % (array.ndim, estimator_name)
1061 )
1063 if force_all_finite:
-> 1064 _assert_all_finite(
1065 array,
1066 input_name=input_name,
1067 estimator_name=estimator_name,
1068 allow_nan=force_all_finite == "allow-nan",
1069 )
1071 if copy:
1072 if _is_numpy_namespace(xp):
1073 # only make a copy if `array` and `array_orig` may share memory`
File ~/opt/anaconda3/envs/cpsc330-24/lib/python3.12/site-packages/sklearn/utils/validation.py:123, in _assert_all_finite(X, allow_nan, msg_dtype, estimator_name, input_name)
120 if first_pass_isfinite:
121 return
--> 123 _assert_all_finite_element_wise(
124 X,
125 xp=xp,
126 allow_nan=allow_nan,
127 msg_dtype=msg_dtype,
128 estimator_name=estimator_name,
129 input_name=input_name,
130 )
File ~/opt/anaconda3/envs/cpsc330-24/lib/python3.12/site-packages/sklearn/utils/validation.py:172, in _assert_all_finite_element_wise(X, xp, allow_nan, msg_dtype, estimator_name, input_name)
155 if estimator_name and input_name == "X" and has_nan_error:
156 # Improve the error message on how to handle missing values in
157 # scikit-learn.
158 msg_err += (
159 f"\n{estimator_name} does not accept missing values"
160 " encoded as NaN natively. For supervised learning, you might want"
(...)
170 "#estimators-that-handle-nan-values"
171 )
--> 172 raise ValueError(msg_err)
ValueError: Input X contains NaN.
KNeighborsRegressor does not accept missing values encoded as NaN natively. For supervised learning, you might want to consider sklearn.ensemble.HistGradientBoostingClassifier and Regressor which accept missing values encoded as NaNs natively. Alternatively, it is possible to preprocess the data, for instance by using an imputer transformer in a pipeline or drop samples with missing values. See https://scikit-learn.org/stable/modules/impute.html You can find a list of all estimators that handle NaN values at the following page: https://scikit-learn.org/stable/modules/impute.html#estimators-that-handle-nan-values
What’s the problem?#
ValueError: Input X contains NaN.
The classifier is not able to deal with missing values (NaNs).
What are possible ways to deal with the problem?
Delete the rows?
Replace them with some reasonable values?
SimpleImputeris a transformer insklearnto deal with this problem. For example,You can impute missing values in categorical columns with the most frequent value.
You can impute the missing values in numeric columns with the mean or median of the column.
X_train.sort_values("bedrooms_per_household")
| longitude | latitude | housing_median_age | households | median_income | rooms_per_household | bedrooms_per_household | population_per_household | |
|---|---|---|---|---|---|---|---|---|
| 20248 | -119.23 | 34.25 | 28.0 | 9.0 | 8.0000 | 2.888889 | 0.333333 | 3.222222 |
| 12649 | -121.47 | 38.51 | 52.0 | 9.0 | 3.6250 | 2.222222 | 0.444444 | 8.222222 |
| 3125 | -117.76 | 35.22 | 4.0 | 6.0 | 1.6250 | 3.000000 | 0.500000 | 1.333333 |
| 12138 | -117.22 | 33.87 | 16.0 | 14.0 | 2.6250 | 4.000000 | 0.500000 | 2.785714 |
| 8219 | -118.21 | 33.79 | 33.0 | 36.0 | 4.5938 | 0.888889 | 0.500000 | 2.666667 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 4591 | -118.28 | 34.06 | 42.0 | 1179.0 | 1.2254 | 2.096692 | NaN | 3.218830 |
| 19485 | -120.98 | 37.66 | 10.0 | 255.0 | 0.9336 | 3.662745 | NaN | 1.572549 |
| 6962 | -118.05 | 33.99 | 38.0 | 357.0 | 3.7328 | 4.535014 | NaN | 2.481793 |
| 14970 | -117.01 | 32.74 | 31.0 | 677.0 | 2.6973 | 5.129985 | NaN | 3.098966 |
| 7763 | -118.10 | 33.91 | 36.0 | 130.0 | 3.6389 | 5.584615 | NaN | 3.769231 |
18576 rows × 8 columns
X_train.shape
X_test.shape
(2064, 8)
imputer = SimpleImputer(strategy="median")
imputer.fit(X_train)
X_train_imp = imputer.transform(X_train)
X_test_imp = imputer.transform(X_test)
Let’s check whether the NaN values have been replaced or not
Note that
imputer.transformreturns annumpyarray and not a dataframe
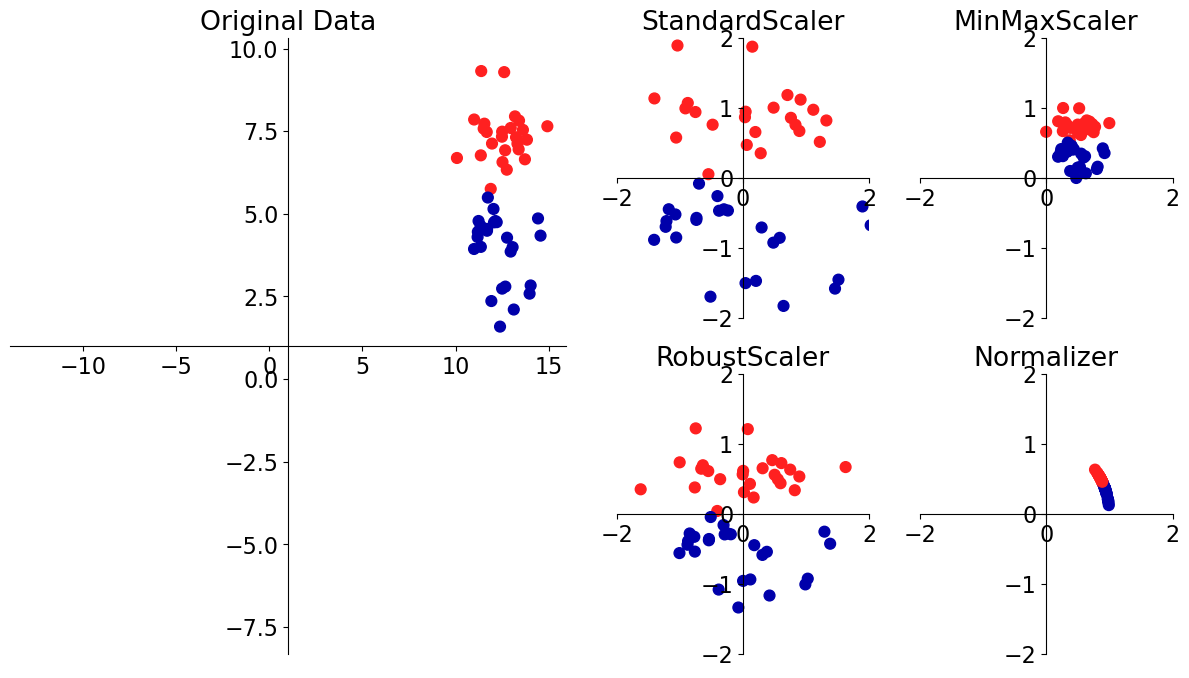
Scaling#
This problem affects a large number of ML methods.
A number of approaches to this problem. We are going to look into two most popular ones.
Approach |
What it does |
How to update \(X\) (but see below!) |
sklearn implementation |
|---|---|---|---|
standardization |
sets sample mean to \(0\), s.d. to \(1\) |
|
There are all sorts of articles on this; see, e.g. here and here.
# [source](https://amueller.github.io/COMS4995-s19/slides/aml-05-preprocessing/#8)
mglearn.plots.plot_scaling()
from sklearn.preprocessing import MinMaxScaler, StandardScaler
scaler = StandardScaler()
X_train_scaled = scaler.fit_transform(X_train_imp) # now on the imputed samples
X_test_scaled = scaler.transform(X_test_imp)
pd.DataFrame(X_train_scaled, columns=X_train.columns)
| longitude | latitude | housing_median_age | households | median_income | rooms_per_household | bedrooms_per_household | population_per_household | |
|---|---|---|---|---|---|---|---|---|
| 0 | 0.908140 | -0.743917 | -0.526078 | 0.266135 | -0.389736 | -0.210591 | -0.083813 | 0.126398 |
| 1 | -0.002057 | 1.083123 | -0.923283 | -1.253312 | -0.198924 | 4.726412 | 11.166631 | -0.050132 |
| 2 | 1.218207 | -1.352930 | 1.380504 | 0.542873 | -0.635239 | -0.273606 | -0.025391 | -0.099240 |
| 3 | 1.128188 | -0.753286 | -0.843842 | -0.561467 | 0.714077 | 0.122307 | -0.280310 | 0.010183 |
| 4 | 1.168196 | -1.287344 | -0.843842 | 2.500924 | -1.059242 | -0.640266 | -0.190617 | 0.126808 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 18571 | 0.733102 | -0.804818 | 0.586095 | -0.966131 | -0.118182 | 0.063110 | -0.099558 | 0.071541 |
| 18572 | 1.163195 | -1.057793 | -1.161606 | 0.728235 | 0.357500 | 0.235096 | -0.163397 | 0.007458 |
| 18573 | -1.097293 | 0.797355 | -1.876574 | 0.514155 | 0.934269 | 0.211892 | -0.135305 | 0.044029 |
| 18574 | -1.437367 | 1.008167 | 1.221622 | -0.454427 | 0.006578 | -0.273382 | -0.149822 | -0.132875 |
| 18575 | 0.242996 | 0.272667 | -0.684960 | -0.396991 | -0.711754 | 0.025998 | 0.042957 | 0.051269 |
18576 rows × 8 columns
knn = KNeighborsRegressor()
knn.fit(X_train_scaled, y_train)
knn.score(X_train_scaled, y_train)
0.7978563117812038
Big difference in the KNN training performance after scaling the data.
But we saw last week that training score doesn’t tell us much. We should look at the cross-validation score.
❓❓ Questions for you#
(iClicker) Exercise 5.2#
iClicker cloud join link: https://join.iclicker.com/VYFJ
Select all of the following statements which are TRUE.
StandardScalerensures a fixed range (i.e., minimum and maximum values) for the features.StandardScalercalculates mean and standard deviation for each feature separately.In general, it’s a good idea to apply scaling on numeric features before training \(k\)-NN or SVM RBF models.
The transformed feature values might be hard to interpret for humans.
After applying
SimpleImputerThe transformed data has a different shape than the original data.
Break (5 min)#

Feature transformations and the golden rule#
How to carry out cross-validation?#
Last week we saw that cross validation is a better way to get a realistic assessment of the model.
Let’s try cross-validation with transformed data.
knn = KNeighborsRegressor()
scaler = StandardScaler()
scaler.fit(X_train_imp)
X_train_scaled = scaler.transform(X_train_imp)
X_test_scaled = scaler.transform(X_test_imp)
scores = cross_validate(knn, X_train_scaled, y_train, return_train_score=True)
pd.DataFrame(scores)
| fit_time | score_time | test_score | train_score | |
|---|---|---|---|---|
| 0 | 0.018903 | 0.146373 | 0.696373 | 0.794236 |
| 1 | 0.008028 | 0.128741 | 0.684447 | 0.791467 |
| 2 | 0.008384 | 0.136767 | 0.695532 | 0.789436 |
| 3 | 0.007950 | 0.139290 | 0.679478 | 0.793243 |
| 4 | 0.007925 | 0.084661 | 0.680657 | 0.794820 |
Do you see any problem here?
Are we applying
fit_transformon train portion andtransformon validation portion in each fold?Here you might be allowing information from the validation set to leak into the training step.
You need to apply the SAME preprocessing steps to train/validation.
With many different transformations and cross validation the code gets unwieldy very quickly.
Likely to make mistakes and “leak” information.
In these examples our test accuracies look fine, but our methodology is flawed.
Implications can be significant in practice!
Pipelines#
Can we do this in a more elegant and organized way?
YES!! Using
scikit-learn Pipeline.scikit-learn Pipelineallows you to define a “pipeline” of transformers with a final estimator.
Let’s combine the preprocessing and model with pipeline
### Simple example of a pipeline
from sklearn.pipeline import Pipeline
pipe = Pipeline(
steps=[
("imputer", SimpleImputer(strategy="median")),
("scaler", StandardScaler()),
("regressor", KNeighborsRegressor()),
]
)
Syntax: pass in a list of steps.
The last step should be a model/classifier/regressor.
All the earlier steps should be transformers.
Alternative and more compact syntax: make_pipeline#
Shorthand for
PipelineconstructorDoes not permit naming steps
Instead the names of steps are set to lowercase of their types automatically;
StandardScaler()would be named asstandardscaler
from sklearn.pipeline import make_pipeline
pipe = make_pipeline(
SimpleImputer(strategy="median"), StandardScaler(), KNeighborsRegressor()
)
pipe.fit(X_train, y_train)
Pipeline(steps=[('simpleimputer', SimpleImputer(strategy='median')),
('standardscaler', StandardScaler()),
('kneighborsregressor', KNeighborsRegressor())])In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook. On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
Pipeline(steps=[('simpleimputer', SimpleImputer(strategy='median')),
('standardscaler', StandardScaler()),
('kneighborsregressor', KNeighborsRegressor())])SimpleImputer(strategy='median')
StandardScaler()
KNeighborsRegressor()
Note that we are passing
X_trainand not the imputed or scaled data here.
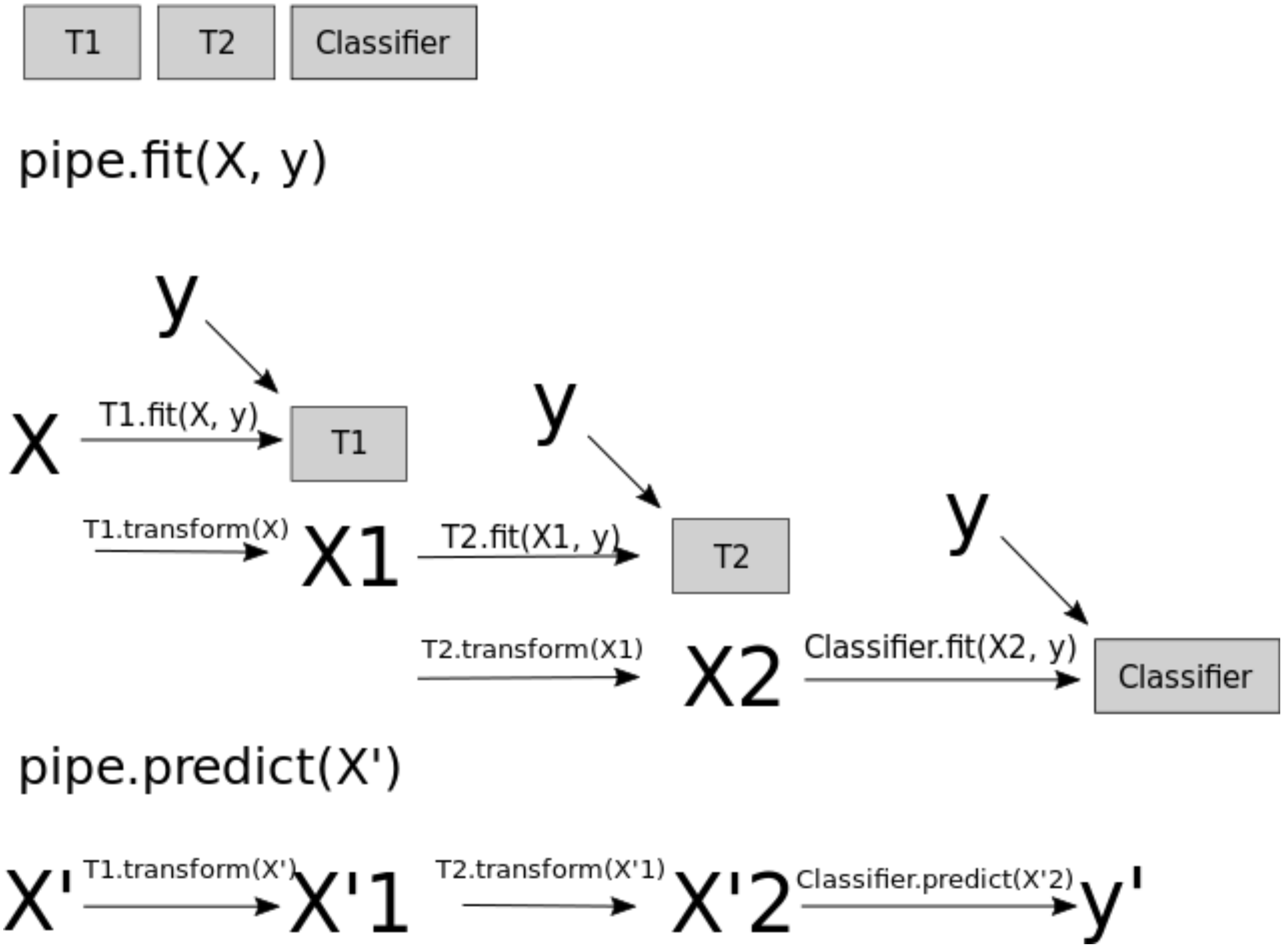
When you call fit on the pipeline, it carries out the following steps:
Fit
SimpleImputeronX_trainTransform
X_trainusing the fitSimpleImputerto createX_train_impFit
StandardScaleronX_train_impTransform
X_train_impusing the fitStandardScalerto createX_train_imp_scaledFit the model (
KNeighborsRegressorin our case) onX_train_imp_scaled
pipe.predict(X_train)
array([126500., 117380., 187700., ..., 259500., 308120., 60860.])
Note that we are passing original data to predict as well. This time the pipeline is carrying out following steps:
Transform
X_trainusing the fitSimpleImputerto createX_train_impTransform
X_train_impusing the fitStandardScalerto createX_train_imp_scaledPredict using the fit model (
KNeighborsRegressorin our case) onX_train_imp_scaled.
Let’s try cross-validation with our pipeline#
results_dict["imp + scaling + knn"] = mean_std_cross_val_scores(
pipe, X_train, y_train, return_train_score=True
)
pd.DataFrame(results_dict).T
/var/folders/7l/2m7m0lw97rvf654x1cwtdfmr0000gr/T/ipykernel_53933/4158382658.py:26: FutureWarning: Series.__getitem__ treating keys as positions is deprecated. In a future version, integer keys will always be treated as labels (consistent with DataFrame behavior). To access a value by position, use `ser.iloc[pos]`
out_col.append((f"%0.3f (+/- %0.3f)" % (mean_scores[i], std_scores[i])))
| fit_time | score_time | test_score | train_score | |
|---|---|---|---|---|
| dummy | 0.003 (+/- 0.003) | 0.004 (+/- 0.006) | -0.055 (+/- 0.012) | -0.055 (+/- 0.001) |
| imp + scaling + knn | 0.025 (+/- 0.013) | 0.134 (+/- 0.013) | 0.693 (+/- 0.014) | 0.797 (+/- 0.015) |
Using a Pipeline takes care of applying the fit_transform on the train portion and only transform on the validation portion in each fold.
Categorical features [video]#
Recall that we had dropped the categorical feature
ocean_proximityfeature from the dataframe. But it could potentially be a useful feature in this task.Let’s create our
X_trainand andX_testagain by keeping the feature in the data.
X_train
| longitude | latitude | housing_median_age | households | median_income | rooms_per_household | bedrooms_per_household | population_per_household | |
|---|---|---|---|---|---|---|---|---|
| 6051 | -117.75 | 34.04 | 22.0 | 602.0 | 3.1250 | 4.897010 | 1.056478 | 4.318937 |
| 20113 | -119.57 | 37.94 | 17.0 | 20.0 | 3.4861 | 17.300000 | 6.500000 | 2.550000 |
| 14289 | -117.13 | 32.74 | 46.0 | 708.0 | 2.6604 | 4.738701 | 1.084746 | 2.057910 |
| 13665 | -117.31 | 34.02 | 18.0 | 285.0 | 5.2139 | 5.733333 | 0.961404 | 3.154386 |
| 14471 | -117.23 | 32.88 | 18.0 | 1458.0 | 1.8580 | 3.817558 | 1.004801 | 4.323045 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 7763 | -118.10 | 33.91 | 36.0 | 130.0 | 3.6389 | 5.584615 | NaN | 3.769231 |
| 15377 | -117.24 | 33.37 | 14.0 | 779.0 | 4.5391 | 6.016688 | 1.017972 | 3.127086 |
| 17730 | -121.76 | 37.33 | 5.0 | 697.0 | 5.6306 | 5.958393 | 1.031564 | 3.493544 |
| 15725 | -122.44 | 37.78 | 44.0 | 326.0 | 3.8750 | 4.739264 | 1.024540 | 1.720859 |
| 19966 | -119.08 | 36.21 | 20.0 | 348.0 | 2.5156 | 5.491379 | 1.117816 | 3.566092 |
18576 rows × 8 columns
X_train = train_df.drop(columns=["median_house_value"])
y_train = train_df["median_house_value"]
X_test = test_df.drop(columns=["median_house_value"])
y_test = test_df["median_house_value"]
Let’s try to build a
KNeighborRegressoron this data using our pipeline
pipe.fit(X_train, X_train)
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
Cell In[51], line 1
----> 1 pipe.fit(X_train, X_train)
File ~/opt/anaconda3/envs/cpsc330-24/lib/python3.12/site-packages/sklearn/base.py:1473, in _fit_context.<locals>.decorator.<locals>.wrapper(estimator, *args, **kwargs)
1466 estimator._validate_params()
1468 with config_context(
1469 skip_parameter_validation=(
1470 prefer_skip_nested_validation or global_skip_validation
1471 )
1472 ):
-> 1473 return fit_method(estimator, *args, **kwargs)
File ~/opt/anaconda3/envs/cpsc330-24/lib/python3.12/site-packages/sklearn/pipeline.py:469, in Pipeline.fit(self, X, y, **params)
426 """Fit the model.
427
428 Fit all the transformers one after the other and sequentially transform the
(...)
466 Pipeline with fitted steps.
467 """
468 routed_params = self._check_method_params(method="fit", props=params)
--> 469 Xt = self._fit(X, y, routed_params)
470 with _print_elapsed_time("Pipeline", self._log_message(len(self.steps) - 1)):
471 if self._final_estimator != "passthrough":
File ~/opt/anaconda3/envs/cpsc330-24/lib/python3.12/site-packages/sklearn/pipeline.py:406, in Pipeline._fit(self, X, y, routed_params)
404 cloned_transformer = clone(transformer)
405 # Fit or load from cache the current transformer
--> 406 X, fitted_transformer = fit_transform_one_cached(
407 cloned_transformer,
408 X,
409 y,
410 None,
411 message_clsname="Pipeline",
412 message=self._log_message(step_idx),
413 params=routed_params[name],
414 )
415 # Replace the transformer of the step with the fitted
416 # transformer. This is necessary when loading the transformer
417 # from the cache.
418 self.steps[step_idx] = (name, fitted_transformer)
File ~/opt/anaconda3/envs/cpsc330-24/lib/python3.12/site-packages/joblib/memory.py:312, in NotMemorizedFunc.__call__(self, *args, **kwargs)
311 def __call__(self, *args, **kwargs):
--> 312 return self.func(*args, **kwargs)
File ~/opt/anaconda3/envs/cpsc330-24/lib/python3.12/site-packages/sklearn/pipeline.py:1310, in _fit_transform_one(transformer, X, y, weight, message_clsname, message, params)
1308 with _print_elapsed_time(message_clsname, message):
1309 if hasattr(transformer, "fit_transform"):
-> 1310 res = transformer.fit_transform(X, y, **params.get("fit_transform", {}))
1311 else:
1312 res = transformer.fit(X, y, **params.get("fit", {})).transform(
1313 X, **params.get("transform", {})
1314 )
File ~/opt/anaconda3/envs/cpsc330-24/lib/python3.12/site-packages/sklearn/utils/_set_output.py:313, in _wrap_method_output.<locals>.wrapped(self, X, *args, **kwargs)
311 @wraps(f)
312 def wrapped(self, X, *args, **kwargs):
--> 313 data_to_wrap = f(self, X, *args, **kwargs)
314 if isinstance(data_to_wrap, tuple):
315 # only wrap the first output for cross decomposition
316 return_tuple = (
317 _wrap_data_with_container(method, data_to_wrap[0], X, self),
318 *data_to_wrap[1:],
319 )
File ~/opt/anaconda3/envs/cpsc330-24/lib/python3.12/site-packages/sklearn/base.py:1101, in TransformerMixin.fit_transform(self, X, y, **fit_params)
1098 return self.fit(X, **fit_params).transform(X)
1099 else:
1100 # fit method of arity 2 (supervised transformation)
-> 1101 return self.fit(X, y, **fit_params).transform(X)
File ~/opt/anaconda3/envs/cpsc330-24/lib/python3.12/site-packages/sklearn/base.py:1473, in _fit_context.<locals>.decorator.<locals>.wrapper(estimator, *args, **kwargs)
1466 estimator._validate_params()
1468 with config_context(
1469 skip_parameter_validation=(
1470 prefer_skip_nested_validation or global_skip_validation
1471 )
1472 ):
-> 1473 return fit_method(estimator, *args, **kwargs)
File ~/opt/anaconda3/envs/cpsc330-24/lib/python3.12/site-packages/sklearn/impute/_base.py:421, in SimpleImputer.fit(self, X, y)
403 @_fit_context(prefer_skip_nested_validation=True)
404 def fit(self, X, y=None):
405 """Fit the imputer on `X`.
406
407 Parameters
(...)
419 Fitted estimator.
420 """
--> 421 X = self._validate_input(X, in_fit=True)
423 # default fill_value is 0 for numerical input and "missing_value"
424 # otherwise
425 if self.fill_value is None:
File ~/opt/anaconda3/envs/cpsc330-24/lib/python3.12/site-packages/sklearn/impute/_base.py:348, in SimpleImputer._validate_input(self, X, in_fit)
342 if "could not convert" in str(ve):
343 new_ve = ValueError(
344 "Cannot use {} strategy with non-numeric data:\n{}".format(
345 self.strategy, ve
346 )
347 )
--> 348 raise new_ve from None
349 else:
350 raise ve
ValueError: Cannot use median strategy with non-numeric data:
could not convert string to float: 'INLAND'
This failed because we have non-numeric data.
Imagine how \(k\)-NN would calculate distances when you have non-numeric features.
Can we use this feature in the model?#
In
scikit-learn, most algorithms require numeric inputs.Decision trees could theoretically work with categorical features.
However, the sklearn implementation does not support this.
What are the options?#
Drop the column (not recommended)
If you know that the column is not relevant to the target in any way you may drop it.
We can transform categorical features to numeric ones so that we can use them in the model.
Ordinal encoding (occasionally recommended)
One-hot encoding (recommended in most cases) (this lecture)
X_toy = pd.DataFrame(
{
"language": [
"English",
"Vietnamese",
"English",
"Mandarin",
"English",
"English",
"Mandarin",
"English",
"Vietnamese",
"Mandarin",
"French",
"Spanish",
"Mandarin",
"Hindi",
]
}
)
X_toy
| language | |
|---|---|
| 0 | English |
| 1 | Vietnamese |
| 2 | English |
| 3 | Mandarin |
| 4 | English |
| 5 | English |
| 6 | Mandarin |
| 7 | English |
| 8 | Vietnamese |
| 9 | Mandarin |
| 10 | French |
| 11 | Spanish |
| 12 | Mandarin |
| 13 | Hindi |
Ordinal encoding (occasionally recommended)#
Here we simply assign an integer to each of our unique categorical labels.
We can use sklearn’s
OrdinalEncoder.
from sklearn.preprocessing import OrdinalEncoder
enc = OrdinalEncoder()
enc.fit(X_toy)
X_toy_ord = enc.transform(X_toy)
df = pd.DataFrame(
data=X_toy_ord,
columns=["language_enc"],
index=X_toy.index,
)
pd.concat([X_toy, df], axis=1)
| language | language_enc | |
|---|---|---|
| 0 | English | 0.0 |
| 1 | Vietnamese | 5.0 |
| 2 | English | 0.0 |
| 3 | Mandarin | 3.0 |
| 4 | English | 0.0 |
| 5 | English | 0.0 |
| 6 | Mandarin | 3.0 |
| 7 | English | 0.0 |
| 8 | Vietnamese | 5.0 |
| 9 | Mandarin | 3.0 |
| 10 | French | 1.0 |
| 11 | Spanish | 4.0 |
| 12 | Mandarin | 3.0 |
| 13 | Hindi | 2.0 |
What’s the problem with this approach?
We have imposed ordinality on the categorical data.
For example, imagine when you are calculating distances. Is it fair to say that French and Hindi are closer than French and Spanish?
In general, label encoding is useful if there is ordinality in your data and capturing it is important for your problem, e.g.,
[cold, warm, hot].
One-hot encoding (OHE)#
Create new binary columns to represent our categories.
If we have \(c\) categories in our column.
We create \(c\) new binary columns to represent those categories.
Example: Imagine a language column which has the information on whether you
We can use sklearn’s
OneHotEncoderto do so.
Note
One-hot encoding is called one-hot because only one of the newly created features is 1 for each data point.
from sklearn.preprocessing import OneHotEncoder
enc = OneHotEncoder(handle_unknown="ignore", sparse_output=False)
enc.fit(X_toy)
X_toy_ohe = enc.transform(X_toy)
pd.DataFrame(
data=X_toy_ohe,
columns=enc.get_feature_names_out(["language"]),
index=X_toy.index,
)
| language_English | language_French | language_Hindi | language_Mandarin | language_Spanish | language_Vietnamese | |
|---|---|---|---|---|---|---|
| 0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 |
| 2 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 3 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 |
| 4 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 5 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 6 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 |
| 7 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 8 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 |
| 9 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 |
| 10 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 11 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 |
| 12 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 |
| 13 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 |
Let’s do it on our housing data#
ohe = OneHotEncoder(sparse_output=False, dtype="int")
ohe.fit(X_train[["ocean_proximity"]])
X_imp_ohe_train = ohe.transform(X_train[["ocean_proximity"]])
We can look at the new features created using
categories_attribute
ohe.categories_
[array(['<1H OCEAN', 'INLAND', 'ISLAND', 'NEAR BAY', 'NEAR OCEAN'],
dtype=object)]
transformed_ohe = pd.DataFrame(
data=X_imp_ohe_train,
columns=ohe.get_feature_names_out(["ocean_proximity"]),
index=X_train.index,
)
transformed_ohe
| ocean_proximity_<1H OCEAN | ocean_proximity_INLAND | ocean_proximity_ISLAND | ocean_proximity_NEAR BAY | ocean_proximity_NEAR OCEAN | |
|---|---|---|---|---|---|
| 6051 | 0 | 1 | 0 | 0 | 0 |
| 20113 | 0 | 1 | 0 | 0 | 0 |
| 14289 | 0 | 0 | 0 | 0 | 1 |
| 13665 | 0 | 1 | 0 | 0 | 0 |
| 14471 | 0 | 0 | 0 | 0 | 1 |
| ... | ... | ... | ... | ... | ... |
| 7763 | 1 | 0 | 0 | 0 | 0 |
| 15377 | 1 | 0 | 0 | 0 | 0 |
| 17730 | 1 | 0 | 0 | 0 | 0 |
| 15725 | 0 | 0 | 0 | 1 | 0 |
| 19966 | 0 | 1 | 0 | 0 | 0 |
18576 rows × 5 columns
See also
One-hot encoded variables are also referred to as dummy variables.
You will often see people using get_dummies method of pandas to convert categorical variables into dummy variables. That said, using sklearn’s OneHotEncoder has the advantage of making it easy to treat training and test set in a consistent way.
❓❓ Questions for you#
(iClicker) Exercise 5.3#
iClicker cloud join link: https://join.iclicker.com/VYFJ
Select all of the following statements which are TRUE.
You can have scaling of numeric features, one-hot encoding of categorical features, and
scikit-learnestimator within a single pipeline.Once you have a
scikit-learnpipeline object with an estimator as the last step, you can callfit,predict, andscoreon it.We have to be careful of the order we put each transformation and model in a pipeline.
If you call
cross_validatewith a pipeline object, it will callfitandtransformon the training fold and onlytransformon the validation fold.
What did we learn today?#
Motivation for preprocessing
Common preprocessing steps
Imputation
Scaling
One-hot encoding
Golden rule in the context of preprocessing
Building simple supervised machine learning pipelines using
sklearn.pipeline.make_pipeline.
Problem: Different transformations on different columns#
How do we put this together with other columns in the data before fitting the regressor?
Before we fit our regressor, we want to apply different transformations on different columns
Numeric columns
imputation
scaling
Categorical columns
imputation
one-hot encoding
Coming up: sklearn’s ColumnTransformer!!
