CPSC 330 Python notes#
About this document#
This document contains some Python lecture materials from the 1st offering of CPSC 330. We have decided to stop allocated lecture time to this topic and instead have this as reference material.
import numpy as np
import pandas as pd
Plotting with matplotlib#
We will use matplotlib as our plotting library.
For those familiar with MATLAB, this package is based on MATLAB plotting.
To use matplotlib, we first import it:
import matplotlib.pyplot as plt
We can now use functions in
pltto plot things:
x = [1,2,3]
y = [4,4,5]
plt.plot(x,y)
[<matplotlib.lines.Line2D at 0x1227a36e0>]
You will often see me put a semicolon at the end of a line.
This is only relevant to Jupyter; it suppresses the line of “output”..
plt.plot(x,y);

In your homework assignments, at a minimum, you should have axis labels for every figure that you submit.
plt.plot(x,y)
plt.xlabel("the independent variable")
plt.ylabel("the dependent variable");
If you are plotting multiple curves, make sure you include a legend!
plt.plot(x,y, label="label is y")
plt.plot(x,x, label="x")
plt.xlabel("the independent variable")
plt.ylabel("the dependent variables")
plt.legend();
You will likely need to visit the matplotlib.pyplot documentation when trying to do other things.
When you save an
.ipynbfile, the output, including plots, is stored in the file.This is a hassle for git.
But it’s also convenient.
This is how you will submit plots.
Numpy arrays#
Basic numpy is covered in the posted videos, you are expected to have a basic knowledge of numpy.
x = np.zeros(4)
x
array([0., 0., 0., 0.])
y = np.ones(4)
x+y
array([1., 1., 1., 1.])
z = np.random.rand(2,3)
z
array([[0.17393037, 0.16991815, 0.18317085],
[0.56521734, 0.37050406, 0.33888659]])
z[0,1]
0.1699181533555938
Numpy array shapes#
One of the most confusing things about numpy: what I call a “1-D array” can have 3 possible shapes:
x = np.ones(5)
print(x)
print("size:", x.size)
print("ndim:", x.ndim)
print("shape:",x.shape)
[1. 1. 1. 1. 1.]
size: 5
ndim: 1
shape: (5,)
y = np.ones((1,5))
print(y)
print("size:", y.size)
print("ndim:", y.ndim)
print("shape:",y.shape)
[[1. 1. 1. 1. 1.]]
size: 5
ndim: 2
shape: (1, 5)
z = np.ones((5,1))
print(z)
print("size:", z.size)
print("ndim:", z.ndim)
print("shape:",z.shape)
[[1.]
[1.]
[1.]
[1.]
[1.]]
size: 5
ndim: 2
shape: (5, 1)
np.array_equal(x,y)
False
np.array_equal(x,z)
False
np.array_equal(y,z)
False
Broadcasting in numpy#
Arrays with different sizes cannot be directly used in arithmetic operations.
Broadcasting describes how numpy treats arrays with different shapes during arithmetic operations.
The idea is to vectorize operations to avoid loops and speed up the code.
Example: I sell pies on the weekends.
I sell 3 types of pies at different prices, and I sold the following number of each pie last weekend.
I want to know how much money I made per pie type per day.

cost = np.array([20, 15, 25])
print("Pie cost:")
print(cost.reshape(3,1))
sales = np.array([[2, 3, 1],
[6, 3, 3],
[5, 3, 5]])
print("\nPie sales (#):")
print(sales)
Pie cost:
[[20]
[15]
[25]]
Pie sales (#):
[[2 3 1]
[6 3 3]
[5 3 5]]
How can we multiply these two arrays together?

Slowest method: nested loop#
total = np.zeros((3, 3))
for i in range(3):
for j in range(3):
total[i,j] = cost[i] * sales[i,j]
total
array([[ 40., 60., 20.],
[ 90., 45., 45.],
[125., 75., 125.]])
Faster method: vectorize the loop over rows#
total = np.zeros((3, 3))
for j in range(3):
total[:,j] = cost * sales[:,j]
total
array([[ 40., 60., 20.],
[ 90., 45., 45.],
[125., 75., 125.]])
No-loop method: make them the same size, and multiply element-wise#

cost_rep = np.repeat(cost[:,np.newaxis], 3, axis=1)
cost_rep
array([[20, 20, 20],
[15, 15, 15],
[25, 25, 25]])
cost_rep * sales
array([[ 40, 60, 20],
[ 90, 45, 45],
[125, 75, 125]])
What is
np.newaxis?It changes the shape:
cost.shape
(3,)
cost[:,np.newaxis].shape
(3, 1)
cost.reshape(3,1).shape # the name thing
(3, 1)
cost[np.newaxis].shape
(1, 3)
Fastest method: broadcasting#
cost[:,np.newaxis] * sales
array([[ 40, 60, 20],
[ 90, 45, 45],
[125, 75, 125]])
numpy does the equivalent of
np.repeat()for you - no need to do it explicitlyIt is debatable whether this code is more readable, but it is definitely faster.
When can we use broadcasting?#
Say we want to broadcast the following two arrays:
arr1 = np.arange(3)
arr2 = np.ones((5))
arr1
array([0, 1, 2])
arr2
array([1., 1., 1., 1., 1.])
arr1.shape
(3,)
arr2.shape
(5,)
The broadcast will fail because the arrays are not compatible…
arr1 + arr2
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
Cell In[33], line 1
----> 1 arr1 + arr2
ValueError: operands could not be broadcast together with shapes (3,) (5,)
We can facilitate this broadcast by adding a dimension using
np.newaxis.np.newaxisincreases the dimension of an array by one dimension.
arr1.shape
arr1 = arr1[:, np.newaxis]
arr1.shape
arr2 = arr2[np.newaxis]
arr2.shape
arr1 + arr2
the opposite, reducing a dimension, can be achieved by
np.squeeze()
arr1.shape
np.squeeze(arr1).shape
The rules of broadcasting:
NumPy compares arrays one dimension at a time. It starts with the trailing dimensions, and works its way to the first dimensions.
dimensions are compatible if:
they are equal, or
one of them is 1.
Use the code below to test out array compatibitlity
a = np.ones((5,1))
b = np.ones((1,3))
print(f"The shape of a is: {a.shape}")
print(f"The shape of b is: {b.shape}")
try:
print(f"The shape of a + b is: {(a + b).shape}")
except:
print(f"ERROR: arrays are NOT broadcast compatible!")
Introduction to pandas#
The most popular Python library for tabular data structures
import pandas as pd
Pandas Series#
A Series is like a NumPy array but with labels
1-dimensional
Can be created from a list, ndarray or dictionary using
pd.Series()Labels may be integers or strings
Here are two series of gold medal counts for the 2012 and 2016 Olympics:
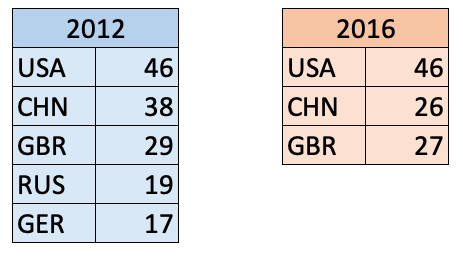
pd.Series()
s1 = pd.Series(data = [46, 38, 29, 19, 17],
index = ['USA','CHN','GBR','RUS','GER'])
s1
s2 = pd.Series([46, 26, 27],
['USA', 'CHN', 'GBR'])
s2
Like ndarrays we use square brackets
[]to index a seriesBUT, Series can be indexed by an integer location OR a label
s1
s1.iloc[0]
s1.iloc[1]
s1["USA"]
s1["USA":"RUS"]
Do we expect these two series to be compatible for broadcasting?
s1
s2
print(f"The shape of s1 is: {s1.shape}")
print(f"The shape of s2 is: {s2.shape}")
s1 + s2
Unlike ndarrays operations between Series (+, -, /, *) align values based on their LABELS
The result index will be the sorted union of the two indexes
Pandas DataFrames#
The primary Pandas data structure
Really just a bunch of Series (with the same index labels) stuck together
Made using
pd.DataFrame()
Creating a DataFrame with a numpy array
d = np.array([[46, 46],
[38, 26],
[29, 27]])
c = ['2012', '2016']
i = ['USA', 'CHN', 'GBR']
df = pd.DataFrame(data=d, index=i, columns=c)
df
(optional) Creating a DataFrame with a dictionary
d = {'2012': [46, 38, 29],
'2016': [46, 26, 27]}
i = ['USA', 'CHN', 'GBR']
df = pd.DataFrame(d, i)
df
Indexing Dataframes#
There are three main ways to index a DataFrame:
[](slice for rows, label for columns).loc[].iloc[]
df
[] notation#
you can index columns by single labels or lists of labels
df['2012']
type(df['2012'])
type(['2012', '2016'])
df[['2012', '2016']]
(optional) you can also index rows with [], but you can only index rows with slices
df["CHN":"GBR"]
# df["USA"] # doesn't work
df[:"USA"] # does work
this is a little unintuitive, so pandas created two other ways to index a dataframe:
for indexing with integers:
df.iloc[]for indexing with labels:
df.loc[]
df
df.iloc[1]
df.iloc[2,1]
df.loc['CHN']
df.loc['GBR', '2016']
df.loc[['USA', 'GBR'], ['2012']]
df.index
df.columns
#df.loc[df.index[0], '2016']
#df.loc['USA', df.columns[0]]
Indexing cheatsheet#
[]accepts slices for row indexing or labels (single or list) for column indexing.iloc[]accepts integers for row/column indexing, and can be single values or lists.loc[]accepts labels for row/column indexing, and can be single values or listsfor integer row/named column:
df.loc[df.index[#], 'labels']for named row/integer column:
df.loc['labels', df.columns[#]]
Break (5 min)#
Reading from .csv#
Most of the time you will be loading .csv files for use in pandas using
pd.read_csv()Example dataset: a colleague’s cycling commute to/from UBC everyday
path = 'data/cycling_data.csv'
pd.read_csv(path, index_col=0, parse_dates=True).head()
Reading from url#
you may also want to read directly from an url at times
pd.read_csv()accepts urls as input
url = 'https://raw.githubusercontent.com/TomasBeuzen/toy-datasets/master/wine_1.csv'
pd.read_csv(url)
Reading from other formats#
pd.read_excel()pd.read_html()pd.read_json()etc
Dataframe summaries#
df = pd.read_csv('data/cycling_data.csv')
df.head()
df.info()
df.describe(include='all')
Renaming columns with df.rename()#
df.head()
we can rename specific columns using
df.rename()
{"Comments": "Notes"}
type({"Comments": "Notes"})
df = df.rename(columns={"Comments": "Notes"})
df.head()
there are two options for making permanent dataframe changes:
set the argument
inplace=True, e.g.,df.rename(..., inplace=True)
re-assign, e.g.,
df = df.rename(...)
df.rename(columns={"Comments": "Notes"}, inplace=True) # inplace
df = df.rename(columns={"Comments": "Notes"}) # re-assign
NOTE:#
the pandas team discourages the use of
inplacefor a few reasonsmostly because not all functions have the argument, hides memory copying, leads to hard-to-find bugs
it is recommend to re-assign (method 2 above)
we can also change all columns at once using a list
df.columns = ['col1', 'col2', 'col3', 'col4', 'col5', 'col6']
df.head()
Adding/removing columns with [] and drop()#
df = pd.read_csv('data/cycling_data.csv')
df.head()
adding a single column
df['Speed'] = 3.14159265358979323
df.head()
dropping a column
df = df.drop(columns="Speed")
df.head()
we can also add/drop multiple columns at a time
df = df.drop(columns=['Type', 'Time'])
df.head()
Adding/removing rows with [] and drop()#
df = pd.read_csv('data/cycling_data.csv')
df.tail()
last_row = df.iloc[-1]
last_row
df.shape
(optional) We can add the row to the end of the dataframe using df.append()
df = df.append(last_row)
df.tail()
df.shape
but now we have the index label
32occurring twice (that can be bad! Why?)
df.loc[32]
df = df.iloc[0:33]
df.tail()
we need can set
ignore_index=Trueto avoid duplicate index labels
df = df.append(last_row, ignore_index=True)
df.tail()
df = df.drop(index=[33])
df.tail()
Sorting a dataframe with df.sort_values()#
df = pd.read_csv('data/cycling_data.csv')
df.head()
df.sort_values(by='Time').head()
df.head()
use the
ascendingargument to specify sort order as ascending or descending
df.sort_values(by="Time", ascending=False).head()
(optional) we can sort by multiple columns in succession by passing in lists
df.sort_values(by=['Name', 'Time'], ascending=[True, False]).head()
we can sort a dataframe back to it’s orginal state (based on index) using
df.sort_index()
df.sort_index().head()
Filtering a dataframe with [] and df.query()#
we’ve already seen how to filter a dataframe using
[],.locand.ilocnotationbut what if we want more control?
df.query()is a powerful tool for filtering data
df = pd.read_csv('data/cycling_data.csv')
df.head()
df.query()accepts a string expression to evaluate, using it’s own syntax
df.query('Time > 2500 and Distance < 13')
df[(df['Time'] > 2500) & (df['Distance'] < 13)]
we can refer to variables in the environment by prefixing them with an
@
thresh = 2800
df.query('Time > @thresh')
Applying functions to a dataframe with df.apply() and df.applymap()#
many common functions are built into Pandas as dataframe methods
e.g.,
df.mean(),df.round(),df.min(),df.max(),df.sum(), etc.
df = pd.read_csv('data/cycling_data.csv')
df.head()
df.mean()
df.min()
df.max()
df.sum()
however there will be times when you want to apply a non-built in function
df.apply()applies a function column-wise or row-wisethe function must be able to operate over an entire row or column at a time
df[['Time', 'Distance']].head()
np.sin(2)
np.sin(0)
you may use functions from other packages, such as numpy
df[['Time', 'Distance']].apply(np.sin).head()
or make your own custom function
df[['Time']].apply(lambda x: x/60).head()
use
df.applymap()for functions that accept and return a scalar
df.info()
float(3)
float([1, 2]) # this function only accepts a single value, so this will fail
df[['Time']].apply(float).head() # fails
df_float_1 = df[['Time']].applymap(float).head() # works with applymap
df_float_1
however, if you’re applying an in-built function, there’s often another (vectorized) way…
from Pandas docs “Note that a vectorized version of func often exists, which will be much faster.”
df_float_2 = df[['Time']].astype(float).head() # alternatively, use astype
df_float_2
# using vectorized .astype
%timeit df[['Time']].astype(float)
# using element-wise .applymap
%timeit df[['Time']].applymap(float)
