Appendix B: Feature Selection#
import os
import sys
sys.path.append(os.path.join(os.path.abspath(".."), "code"))
import IPython
import matplotlib.pyplot as plt
import mglearn
import numpy as np
import pandas as pd
from IPython.display import HTML, display
from plotting_functions import *
from sklearn.dummy import DummyClassifier
from sklearn.linear_model import LogisticRegression
from sklearn.model_selection import cross_val_score, cross_validate, train_test_split
from sklearn.pipeline import Pipeline, make_pipeline
from sklearn.preprocessing import StandardScaler
from sklearn.metrics import ConfusionMatrixDisplay # Recommended method in sklearn 1.0
%matplotlib inline
pd.set_option("display.max_colwidth", 200)
from IPython.display import Image
pd.set_option("display.max_colwidth", 200)
DATA_DIR = "../data/"
(Optional) Search and score#
Define a scoring function \(f(S)\) that measures the quality of the set of features \(S\).
Now search for the set of features \(S\) with the best score.
General idea of search and score methods#
Example: Suppose you have three features: \(A, B, C\)
Compute score for \(S = \{\}\)
Compute score for \(S = \{A\}\)
Compute score for \(S= \{B\}\)
Compute score for \(S = \{C\}\)
Compute score for \(S = \{A,B\}\)
Compute score for \(S = \{A,C\}\)
Compute score for \(S = \{B,C\}\)
Compute score for \(S = \{A,B,C\}\)
Return \(S\) with the best score.
How many distinct combinations we have to try out?
(Optional) Forward or backward selection#
Also called wrapper methods
Shrink or grow feature set by removing or adding one feature at a time
Makes the decision based on whether adding/removing the feature improves the CV score or not

# from sklearn.feature_selection import SequentialFeatureSelector
# pipe_forward = make_pipeline(
# StandardScaler(),
# SequentialFeatureSelector(LogisticRegression(max_iter=1000),
# direction="forward",
# n_features_to_select='auto',
# tol=None),
# RandomForestClassifier(n_estimators=100, random_state=42),
# )
# pd.DataFrame(
# cross_validate(pipe_forward, X_train, y_train, return_train_score=True)
# ).mean()
# pipe_forward = make_pipeline(
# StandardScaler(),
# SequentialFeatureSelector(
# LogisticRegression(max_iter=1000),
# direction="backward",
# n_features_to_select=15),
# RandomForestClassifier(n_estimators=100, random_state=42),
# )
# pd.DataFrame(
# cross_validate(pipe_forward, X_train, y_train, return_train_score=True)
# ).mean()
Other ways to search#
Stochastic local search
Inject randomness so that we can explore new parts of the search space
Simulated annealing
Genetic algorithms
Warnings about feature selection#
A feature’s relevance is only defined in the context of other features.
Adding/removing features can make features relevant/irrelevant.
If features can be predicted from other features, you cannot know which one to pick.
Relevance for features does not have a causal relationship.
Don’t be overconfident.
The methods we have seen probably do not discover the ground truth and how the world really works.
They simply tell you which features help in predicting \(y_i\) for the data you have.
(Optional) Problems with feature selection#
The term ‘relevance’ is not clearly defined.
What all things can go wrong with feature selection?
Attribution: From CPSC 340.
Example: Is “Relevance” clearly defined?#
Consider a supervised classification task of predicting whether someone has particular genetic variation (SNP)
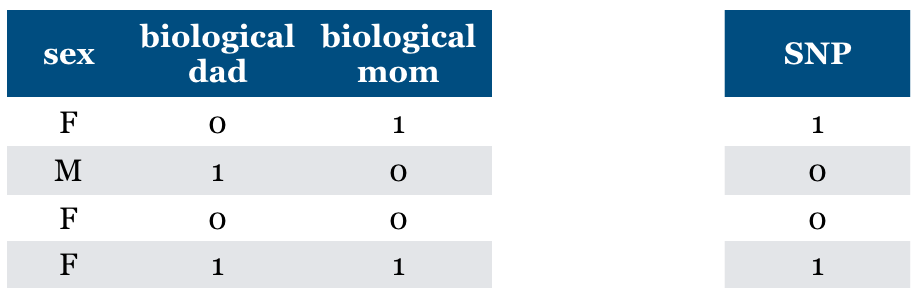
True model: You almost have the same value as your biological mom.
Is “Relevance” clearly defined?#
True model: You almost have the same value for SNP as your biological mom.
(SNP = biological mom) with very high probability
(SNP != biological mom) with very low probability
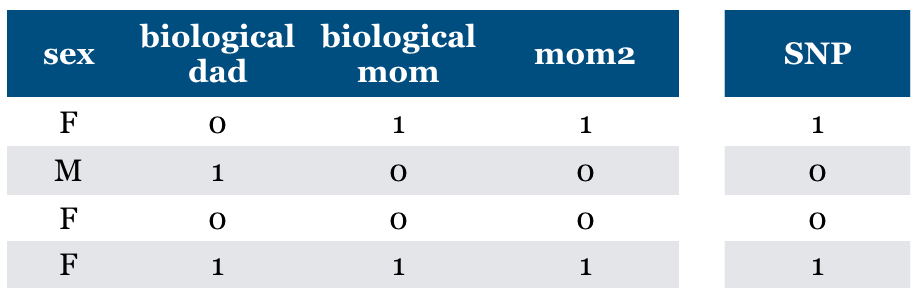
Is “Relevance” clearly defined?#
What if “mom” feature is repeated?
Should we pick both? Should we pick one of them because it predicts the other?
Dependence, collinearity for linear models
If a feature can be predicted from the other, don’t know which one to pick.
Is “Relevance” clearly defined?#
What if we add (maternal) “grandma” feature?
Is it relevant?
We can predict SNP accurately using this feature
Conditional independence
But grandma is irrelevant given biological mom feature
Relevant features may become irrelevant given other features
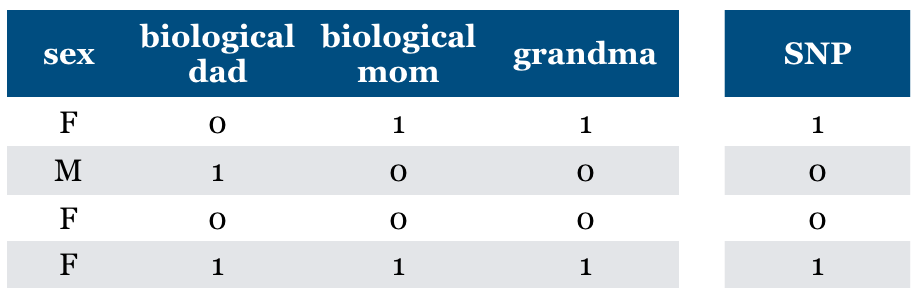
Is “Relevance” clearly defined?#
What if we do not know biological mom feature and we just have grandma feature
It becomes relevant now.
Without mom feature this is the best we can do.
General problem (“taco Tuesday” problem)
Features can become relevant due to missing information
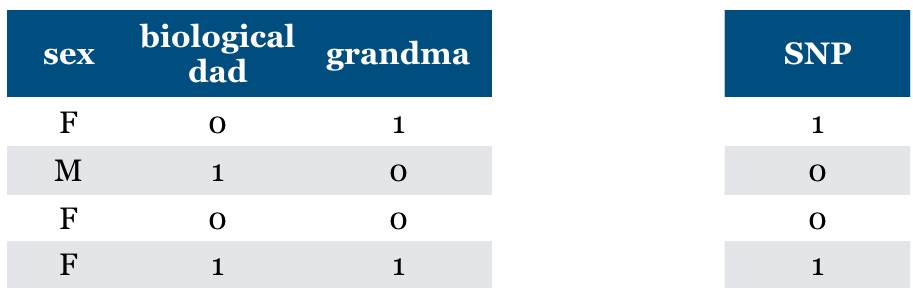
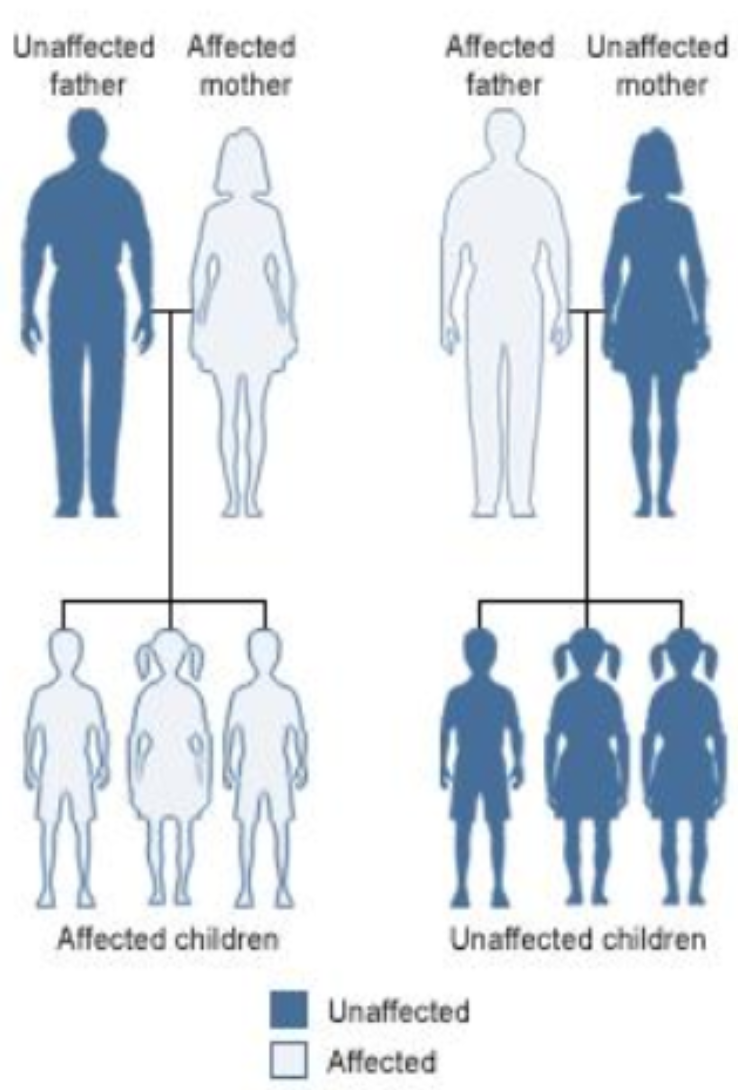
Is “Relevance” clearly defined?#
Are there any relevant features now?
They may have some common maternal ancestor.
What if mom likes dad because they share SNP?
General problem (Confounding)
Hidden features can make irrelevant features relevant.

Is “Relevance” clearly defined?#
Now what if we have “sibling” feature?
The feature is relevant in predicting SNP but not the cause of SNP.
General problem (non causality)
the relevant feature may not be causal
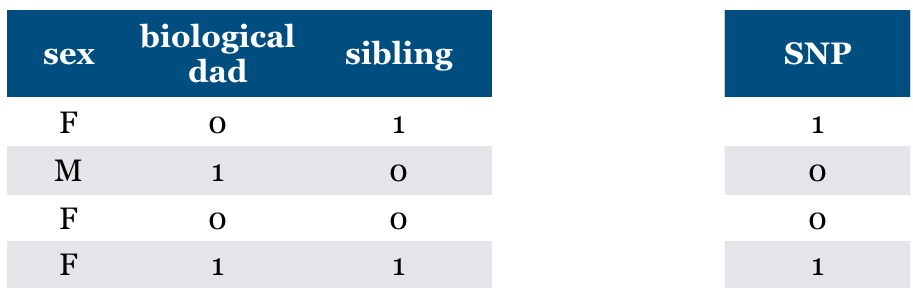
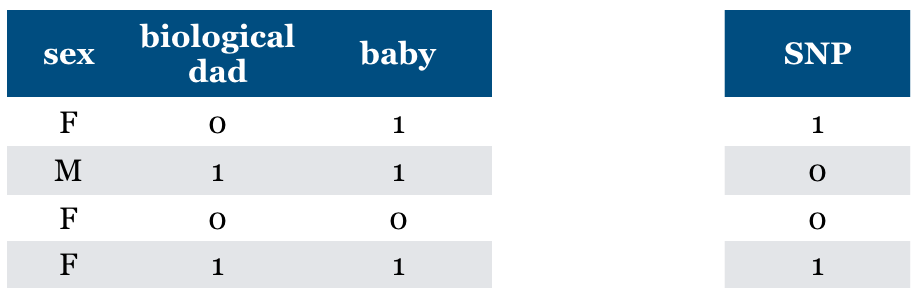
Is “Relevance” clearly defined?#
What if you are given “baby” feature?
Now the sex feature becomes relevant.
“baby” feature is relevant when sex == F
General problem (context specific relevance)
adding a feature can make an irrelevant feature relevant